Putting CENP-A in its place
- PMID: 22729156
- PMCID: PMC4084702
- DOI: 10.1007/s00018-012-1048-8
Putting CENP-A in its place
Abstract
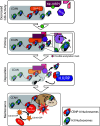
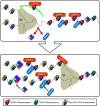
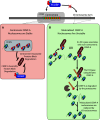
The centromere is the chromosomal region that directs kinetochore assembly during mitosis in order to facilitate the faithful segregation of sister chromatids. The location of the human centromere is epigenetically specified. The presence of nucleosomes that contain the histone H3 variant, CENP-A, are thought to be the epigenetic mark that indicates active centromeres. Maintenance of centromeric identity requires the deposition of new CENP-A nucleosomes with each cell cycle. During S-phase, existing CENP-A nucleosomes are divided among the daughter chromosomes, while new CENP-A nucleosomes are deposited during early G1. The specific assembly of CENP-A nucleosomes at centromeres requires the Mis18 complex, which recruits the CENP-A assembly factor, HJURP. We will review the unique features of centromeric chromatin as well as the mechanism of CENP-A nucleosome deposition. We will also highlight a few recent discoveries that begin to elucidate the factors that temporally and spatially control CENP-A deposition.
Figures



References
-
- Colnaghi R, Carpenter G, Volker M, O’Driscoll M. The consequences of structural genomic alterations in humans: genomic disorders, genomic instability and cancer. Semin Cell Dev Biol. 2011;22:875–885. - PubMed
-
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

