Characterization of the two Neurospora crassa cellobiose dehydrogenases and their connection to oxidative cellulose degradation
- PMID: 22729546
- PMCID: PMC3416632
- DOI: 10.1128/AEM.01503-12
Characterization of the two Neurospora crassa cellobiose dehydrogenases and their connection to oxidative cellulose degradation
Abstract
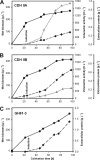
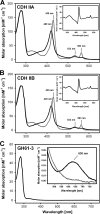
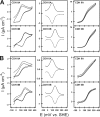
The genome of Neurospora crassa encodes two different cellobiose dehydrogenases (CDHs) with a sequence identity of only 53%. So far, only CDH IIA, which is induced during growth on cellulose and features a C-terminal carbohydrate binding module (CBM), was detected in the secretome of N. crassa and preliminarily characterized. CDH IIB is not significantly upregulated during growth on cellulosic material and lacks a CBM. Since CDH IIB could not be identified in the secretome, both CDHs were recombinantly produced in Pichia pastoris. With the cytochrome domain-dependent one-electron acceptor cytochrome c, CDH IIA has a narrower and more acidic pH optimum than CDH IIB. Interestingly, the catalytic efficiencies of both CDHs for carbohydrates are rather similar, but CDH IIA exhibits 4- to 5-times-higher apparent catalytic constants (k(cat) and K(m) values) than CDH IIB for most tested carbohydrates. A third major difference is the 65-mV-lower redox potential of the heme b cofactor in the cytochrome domain of CDH IIA than CDH IIB. To study the interaction with a member of the glycoside hydrolase 61 family, the copper-dependent polysaccharide monooxygenase GH61-3 (NCU02916) from N. crassa was expressed in P. pastoris. A pH-dependent electron transfer from both CDHs via their cytochrome domains to GH61-3 was observed. The different properties of CDH IIA and CDH IIB and their effect on interactions with GH61-3 are discussed in regard to the proposed in vivo function of the CDH/GH61 enzyme system in oxidative cellulose hydrolysis.
Figures


References
-
- Beeson WT, Phillips CM, Cate JHD, Marletta MA. 2012. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 134: 890–892 - PubMed
-
- Cameron MD, Aust SD. 2001. Cellobiose dehydrogenase—an extracellular fungal flavocytochrome. Enzyme Microb. Technol. 28: 129–138 - PubMed
-
- Coman V, Harreither W, Ludwig R, Haltrich D, Gorton L. 2007. Investigation of electron transfer between cellobiose dehydrogenase from Myriococcum thermophilum and gold electrodes. Chem. Analityczna 52: 945–960
-
- Fan Z, et al. 2012. A novel biochemical route for fuels and chemicals production from cellulosic biomass. PLoS One 7: e31693 doi: 10.1371/journal.pone.0031693 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

