Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism
- PMID: 22729863
- PMCID: PMC3710587
- DOI: 10.1007/978-1-4614-3573-0_12
Bigenomic regulation of cytochrome c oxidase in neurons and the tight coupling between neuronal activity and energy metabolism
Abstract
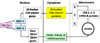
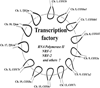
Cytochrome c oxidase is the terminal enzyme of the mitochondrial electron transport chain, without which oxidative metabolism cannot be carried to completion. It is one of only four unique, bigenomic proteins in mammalian cells. The holoenzyme is made up of three mitochondrial-encoded and ten nuclear-encoded subunits in a 1:1 stoichiometry. The ten nuclear subunit genes are located in nine different chromosomes. The coordinated regulation of such a multisubunit, multichromosomal, bigenomic enzyme poses a challenge. It is especially so for neurons, whose mitochondria are widely distributed in extensive dendritic and axonal processes, resulting in the separation of the mitochondrial from the nuclear genome by great distances. Neuronal activity dictates COX activity that reflects protein amount, which, in turn, is regulated at the transcriptional level. All 13 COX transcripts are up- and downregulated by neuronal activity. The ten nuclear COX transcripts and those for Tfam and Tfbms important for mitochondrial COX transcripts are transcribed in the same transcription factory. Bigenomic regulation of all 13 transcripts is mediated by nuclear respiratory factors 1 and 2 (NRF-1 and NRF-2). NRF-1, in addition, also regulates critical neurochemicals of glutamatergic synaptic transmission, thereby ensuring the tight coupling of energy metabolism and neuronal activity at the molecular level in neurons.
Figures

References
-
- Attwell D, Laughlin SB. An energy budget for signaling in the grey matter of the brain. J Cereb Blood Flow Metab. 2001;21:1133–1145. - PubMed
-
- Au HC, Scheffler IE. Promoter analysis of the human succinate dehydrogenase iron-protein gene – both nuclear respiratory factors NRF-1 and NRF-2 are required. Eur J Biochem. 1998;251:164–174. - PubMed
-
- Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, Kelly DP, Holloszy JO. Adaptations of skeletal muscle to exercise: Rapid increase in the transcriptional coactivator PGC-1. Faseb J. 2002;16:1879–1886. - PubMed
-
- Babb TL, Mikuni N, Najm I, Wylie C, Olive M, Dollar C, MacLennan H. Pre- and postnatal expression of NMDA receptors 1 and 2B subunit proteins in the normal rat cortex. Epilepsy Res. 2005;64:23–30. - PubMed
-
- Bachman NJ, Yang TL, Dasen JS, Ernst RE, Lomax MI. Phylogenetic footprinting of the human cytochrome c oxidase subunit VB promoter. Arch Biochem Biophys. 1996;333:152–162. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

