Borrelia burgdorferi cp32 BpaB modulates expression of the prophage NucP nuclease and SsbP single-stranded DNA-binding protein
- PMID: 22730122
- PMCID: PMC3415502
- DOI: 10.1128/JB.00661-12
Borrelia burgdorferi cp32 BpaB modulates expression of the prophage NucP nuclease and SsbP single-stranded DNA-binding protein
Erratum in
-
Correction for Chenail et al., "Borrelia burgdorferi cp32 BpaB Modulates Expression of the Prophage NucP Nuclease and SsbP Single-Stranded DNA-Binding Protein".J Bacteriol. 2017 Sep 5;199(19):e00400-17. doi: 10.1128/JB.00400-17. Print 2017 Oct 1. J Bacteriol. 2017. PMID: 28874457 Free PMC article. No abstract available.
Abstract
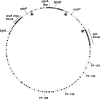
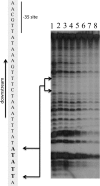
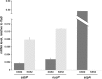
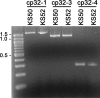
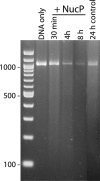
The Borrelia burgdorferi BpaB proteins of the spirochete's ubiquitous cp32 prophages are DNA-binding proteins, required both for maintenance of the bacteriophage episomes and for transcriptional regulation of the cp32 erp operons. Through use of DNase I footprinting, we demonstrate that BpaB binds the erp operator initially at the sequence 5'-TTATA-3'. Electrophoretic mobility shift assays indicated that BpaB also binds with high affinity to sites located in the 5' noncoding regions of two additional cp32 genes. Characterization of the proteins encoded by those genes indicated that they are a single-stranded DNA-binding protein and a nuclease, which we named SsbP and NucP, respectively. Chromatin immunoprecipitation indicated that BpaB binds erp, ssbP, and nucP in live B. burgdorferi. A mutant bacterium that overexpressed BpaB produced significantly higher levels of ssbP and nucP transcript than did the wild-type parent.
Figures



References
-
- Alitalo A, et al. 2002. Complement inhibitor factor H binding to Lyme disease spirochetes is mediated by inducible expression of multiple plasmid-encoded outer surface protein E paralogs. J. Immunol. 169:3847–3853 - PubMed
-
- Beaurepaire C, Chaconas G. 2005. Mapping of essential replication functions of the linear plasmid lp17 of B. burgdorferi by targeted deletion walking. Mol. Microbiol. 57:132–142 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

