The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone
- PMID: 22730123
- PMCID: PMC3415498
- DOI: 10.1128/JB.00830-12
The cryptic competence pathway in Streptococcus pyogenes is controlled by a peptide pheromone
Abstract
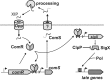
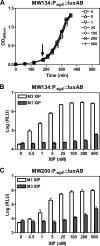
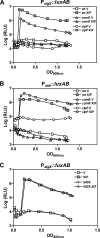
Horizontal gene transfer is an important means of bacterial evolution that is facilitated by transduction, conjugation, and natural genetic transformation. Transformation occurs after bacterial cells enter a state of competence, where naked DNA is acquired from the extracellular environment. Induction of the competent state relies on signals that activate master regulators, causing the expression of genes involved in DNA uptake, processing, and recombination. All streptococcal species contain the master regulator SigX and SigX-dependent effector genes required for natural genetic transformation; however, not all streptococcal species have been shown to be naturally competent. We recently demonstrated that competence development in Streptococcus mutans requires the type II ComRS quorum-sensing circuit, comprising an Rgg transcriptional activator and a novel peptide pheromone (L. Mashburn-Warren, D. A. Morrison, and M. J. Federle, Mol. Microbiol. 78:589-606, 2010). The type II ComRS system is shared by the pyogenic, mutans, and bovis streptococci, including the clinically relevant pathogen Streptococcus pyogenes. Here, we describe the activation of sigX by a small-peptide pheromone and an Rgg regulator of the type II ComRS class. We confirm previous reports that SigX is functional and able to activate sigX-dependent gene expression within the competence regulon, and that SigX stability is influenced by the cytoplasmic protease ClpP. Genomic analyses of available S. pyogenes genomes revealed the presence of intact genes within the competence regulon. While this is the first report to show natural induction of sigX, S. pyogenes remained nontransformable under laboratory conditions. Using radiolabeled DNA, we demonstrate that transformation is blocked at the stage of DNA uptake.
Figures

Similar articles
-
A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator.Mol Microbiol. 2010 Nov;78(3):589-606. doi: 10.1111/j.1365-2958.2010.07361.x. Epub 2010 Sep 14. Mol Microbiol. 2010. PMID: 20969646 Free PMC article.
-
The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in Streptococcus mutans.PLoS Genet. 2015 Jul 9;11(7):e1005353. doi: 10.1371/journal.pgen.1005353. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26158727 Free PMC article.
-
Competence for natural genetic transformation in the Streptococcus bovis group streptococci S. infantarius and S. macedonicus.J Bacteriol. 2013 Jun;195(11):2612-20. doi: 10.1128/JB.00230-13. Epub 2013 Mar 29. J Bacteriol. 2013. PMID: 23543718 Free PMC article.
-
Regulation of competence for natural transformation in streptococci.Infect Genet Evol. 2015 Jul;33:343-60. doi: 10.1016/j.meegid.2014.09.010. Epub 2014 Sep 16. Infect Genet Evol. 2015. PMID: 25236918 Review.
-
Expanding the Vocabulary of Peptide Signals in Streptococcus mutans.Front Cell Infect Microbiol. 2019 Jun 6;9:194. doi: 10.3389/fcimb.2019.00194. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31245303 Free PMC article. Review.
Cited by
-
Bacterial Cell-Cell Communication in the Host via RRNPP Peptide-Binding Regulators.Front Microbiol. 2016 May 20;7:706. doi: 10.3389/fmicb.2016.00706. eCollection 2016. Front Microbiol. 2016. PMID: 27242728 Free PMC article. Review.
-
Interspecies recombination, not de novo mutation, maintains virulence after β-lactam resistance acquisition in Streptococcus pneumoniae.Cell Rep. 2022 Dec 13;41(11):111835. doi: 10.1016/j.celrep.2022.111835. Cell Rep. 2022. PMID: 36516783 Free PMC article.
-
Intensive targeting of regulatory competence genes by transposable elements in streptococci.Mol Genet Genomics. 2019 Jun;294(3):531-548. doi: 10.1007/s00438-018-1507-5. Epub 2018 Nov 7. Mol Genet Genomics. 2019. PMID: 30406402
-
Natural DNA Transformation Is Functional in Lactococcus lactis subsp. cremoris KW2.Appl Environ Microbiol. 2017 Aug 1;83(16):e01074-17. doi: 10.1128/AEM.01074-17. Print 2017 Aug 15. Appl Environ Microbiol. 2017. PMID: 28625996 Free PMC article.
-
A novel chemical inducer of Streptococcus quorum sensing acts by inhibiting the pheromone-degrading endopeptidase PepO.J Biol Chem. 2018 Jan 19;293(3):931-940. doi: 10.1074/jbc.M117.810994. Epub 2017 Dec 4. J Biol Chem. 2018. PMID: 29203527 Free PMC article.
References
-
- Belotserkovsky I, et al. 2009. Functional analysis of the quorum-sensing streptococcal invasion locus (sil). PLoS Pathog. 5:e1000651 doi:10.1371/journal.ppat.1000651 - DOI - PMC - PubMed
-
- Berger-Bachi B. 2002. Resistance mechanisms of gram-positive bacteria. Int. J. Med. Microbiol. 292:27–35 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

