RefZ facilitates the switch from medial to polar division during spore formation in Bacillus subtilis
- PMID: 22730127
- PMCID: PMC3415491
- DOI: 10.1128/JB.00378-12
RefZ facilitates the switch from medial to polar division during spore formation in Bacillus subtilis
Abstract
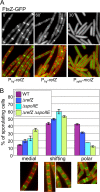
During sporulation, Bacillus subtilis redeploys the division protein FtsZ from midcell to the cell poles, ultimately generating an asymmetric septum. Here, we describe a sporulation-induced protein, RefZ, that facilitates the switch from a medial to a polar FtsZ ring placement. The artificial expression of RefZ during vegetative growth converts FtsZ rings into FtsZ spirals, arcs, and foci, leading to filamentation and lysis. Mutations in FtsZ specifically suppress RefZ-dependent division inhibition, suggesting that RefZ may target FtsZ. During sporulation, cells lacking RefZ are delayed in polar FtsZ ring formation, spending more time in the medial and transition stages of FtsZ ring assembly. A RefZ-green fluorescent protein (GFP) fusion localizes in weak polar foci at the onset of sporulation and as a brighter midcell focus at the time of polar division. RefZ has a TetR DNA binding motif, and point mutations in the putative recognition helix disrupt focus formation and abrogate cell division inhibition. Finally, chromatin immunoprecipitation assays identified sites of RefZ enrichment in the origin region and near the terminus. Collectively, these data support a model in which RefZ helps promote the switch from medial to polar division and is guided by the organization of the chromosome. Models in which RefZ acts as an activator of FtsZ ring assembly near the cell poles or as an inhibitor of the transient medial ring at midcell are discussed.
Figures



References
-
- Adams DW, Errington J. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642–653 - PubMed
-
- Aparicio O, Geisberg JV, Struhl K. 2004. Current protocols in molecular biology. Wiley, New York, NY
-
- Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36 - PubMed
-
- Baumeister R, Helbl V, Hillen W. 1992. Contacts between Tet repressor and tet operator revealed by new recognition specificities of single amino acid replacement mutants. J. Mol. Biol. 226:1257–1270 - PubMed
-
- Baumeister R, Muller G, Hecht B, Hillen W. 1992. Functional roles of amino acid residues involved in forming the alpha-helix-turn-alpha-helix operator DNA binding motif of Tet repressor from Tn10. Proteins 14:168–177 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

