The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance
- PMID: 22730129
- PMCID: PMC3430307
- DOI: 10.1128/JB.00765-12
The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance
Abstract
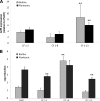
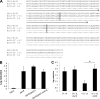
Biofilms are composed of surface-attached microbial communities. A hallmark of biofilms is their profound tolerance of antimicrobial agents. While biofilm drug tolerance has been considered to be multifactorial, our findings indicate, instead, that bacteria within biofilms employ a classical regulatory mechanism to resist the action of antimicrobial agents. Here we report that the transcriptional regulator BrlR, a member of the MerR family of multidrug transport activators, plays a role in the high-level drug tolerance of biofilms formed by Pseudomonas aeruginosa. Expression of brlR was found to be biofilm specific, with brlR inactivation not affecting biofilm formation, motility, or pslA expression but increasing ndvB expression. Inactivation of brlR rendered biofilms but not planktonic cells grown to exponential or stationary phase significantly more susceptible to hydrogen peroxide and five different classes of antibiotics by affecting the MICs and the recalcitrance of biofilms to killing by microbicidal antimicrobial agents. In contrast, overexpression of brlR rendered both biofilms and planktonic cells more tolerant to the same compounds. brlR expression in three cystic fibrosis (CF) isolates was elevated regardless of the mode of growth, suggesting a selection for constitutive brlR expression upon in vivo biofilm formation associated with chronic infections. Despite increased brlR expression, however, isolate CF1-8 was as susceptible to tobramycin as was a ΔbrlR mutant because of a nonsense mutation in brlR. Our results indicate for the first time that biofilms employ a specific regulatory mechanism to resist the action of antimicrobial agents in a BrlR-dependent manner which affects MIC and recalcitrance to killing by microbicidal antimicrobial agents.
Figures

Comment in
-
Regulating antibiotic tolerance within biofilm microcolonies.J Bacteriol. 2012 Sep;194(18):4791-2. doi: 10.1128/JB.01187-12. Epub 2012 Jul 13. J Bacteriol. 2012. PMID: 22797753 Free PMC article. No abstract available.
References
-
- Ahmed M, Borsch CM, Taylor SS, Vazquez-Laslop N, Neyfakh AA. 1994. A protein that activates expression of a multidrug efflux transporter upon binding the transporter substrates. J. Biol. Chem. 269:28506–28513 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

