O-linked β-N-acetylglucosaminylation (O-GlcNAcylation) in primary and metastatic colorectal cancer clones and effect of N-acetyl-β-D-glucosaminidase silencing on cell phenotype and transcriptome
- PMID: 22730328
- PMCID: PMC3436545
- DOI: 10.1074/jbc.M112.345546
O-linked β-N-acetylglucosaminylation (O-GlcNAcylation) in primary and metastatic colorectal cancer clones and effect of N-acetyl-β-D-glucosaminidase silencing on cell phenotype and transcriptome
Abstract
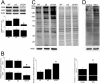
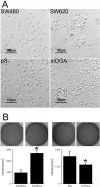
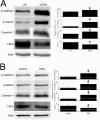
O-linked β-N-acetylglucosamine (O-GlcNAc) glycosylation is a regulatory post-translational modification occurring on the serine or threonine residues of nucleocytoplasmic proteins. O-GlcNAcylation is dynamically regulated by O-GlcNAc transferase and O-GlcNAcase (OGA), which are responsible for O-GlcNAc addition and removal, respectively. Although O-GlcNAcylation was found to play a significant role in several pathologies such as type II diabetes and neurodegenerative diseases, the role of O-GlcNAcylation in the etiology and progression of cancer remains vague. Here, we followed O-GlcNAcylation and its catalytic machinery in metastatic clones of human colorectal cancer and the effect of OGA knockdown on cellular phenotype and on the transcriptome. The colorectal cancer SW620 metastatic clone exhibited increased O-GlcNAcylation and decreased OGA expression compared with its primary clone, SW480. O-GlcNAcylation elevation in SW620 cells, through RNA interference of OGA, resulted in phenotypic alterations that included acquisition of a fibroblast-like morphology, which coincides with epithelial metastatic progression and growth retardation. Microarray analysis revealed that OGA silencing altered the expression of about 1300 genes, mostly involved in cell movement and growth, and specifically affected metabolic pathways of lipids and carbohydrates. These findings support the involvement of O-GlcNAcylation in various aspects of tumor cell physiology and suggest that this modification may serve as a link between metabolic changes and cancer.
Figures




References
-
- Zachara N. E., Hart G. W. (2002) The emerging significance of O-GlcNAc in cellular regulation. Chem. Rev. 102, 431–438 - PubMed
-
- Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., Hart G. W. (2004) Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress. A survival response of mammalian cells. J. Biol. Chem. 279, 30133–30142 - PubMed
-
- Hart G. W. (1997) Dynamic O-linked glycosylation of nuclear and cytoskeletal proteins. Annu. Rev. Biochem. 66, 315–335 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

