A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis
- PMID: 22730405
- PMCID: PMC3406912
- DOI: 10.1105/tpc.112.100107
A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis
Abstract
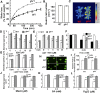
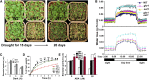
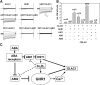
The plant hormone abscisic acid (ABA) regulates stomatal movement under drought stress, and this regulation requires hydrogen peroxide (H2O2). We isolated GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 (GHR1), which encodes a receptor-like kinase localized on the plasma membrane in Arabidopsis thaliana. ghr1 mutants were defective ABA and H2O2 induction of stomatal closure. Genetic analysis indicates that GHR1 is a critical early component in ABA signaling. The ghr1 mutation impaired ABA- and H2O2-regulated activation of S-type anion currents in guard cells. Furthermore, GHR1 physically interacted with, phosphorylated, and activated the S-type anion channel SLOW ANION CHANNEL-ASSOCIATED1 when coexpressed in Xenopus laevis oocytes, and this activation was inhibited by ABA-INSENSITIVE2 (ABI2) but not ABI1. Our study identifies a critical component in ABA and H2O2 signaling that is involved in stomatal movement and resolves a long-standing mystery about the differential functions of ABI1 and ABI2 in this process.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

