Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis
- PMID: 22730461
- PMCID: PMC3396469
- DOI: 10.1073/pnas.1200853109
Genetic and functional analyses implicate the NUDT11, HNF1B, and SLC22A3 genes in prostate cancer pathogenesis
Abstract
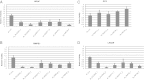
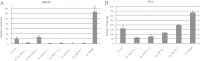
One of the central goals of human genetics is to discover the genes and pathways driving human traits. To date, most of the common risk alleles discovered through genome-wide association studies (GWAS) map to nonprotein-coding regions. Because of our relatively poorer understanding of this part of the genome, the functional consequences of trait-associated variants pose a considerable challenge. To identify the genes through which risk loci act, we hypothesized that the risk variants are regulatory elements. For each of 12 known risk polymorphisms, we evaluated the correlation between risk allele status and transcript abundance for all annotated protein-coding transcripts within a 1-Mb interval. A total of 103 transcripts were evaluated in 662 prostate tissue samples [normal (n = 407) and tumor (n = 255)] from 483 individuals [European Americans (n = 233), Japanese (n = 127), and African Americans (n = 123)]. In a pooled analysis, 4 of the 12 risk variants were strongly associated with five transcripts (NUDT11, MSMB, NCOA4, SLC22A3, and HNF1B) in histologically normal tissue (P ≤ 0.001). Although associations were also observed in tumor tissue, they tended to be more attenuated. Previously, we showed that MSMB and NCOA4 participate in prostate cancer pathogenesis. Suppressing the expression of NUDT11, SLC22A3, and HNF1B influences cellular phenotypes associated with tumor-related properties in prostate cancer cells. Taken together, the data suggest that these transcripts contribute to prostate cancer pathogenesis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Eeles RA, et al. UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology UK ProtecT Study Collaborators PRACTICAL Consortium Identification of seven new prostate cancer susceptibility loci through a genome-wide association study. Nat Genet. 2009;41:1116–1121. - PMC - PubMed
-
- Eeles RA, et al. UK Genetic Prostate Cancer Study Collaborators British Association of Urological Surgeons’ Section of Oncology UK ProtecT Study Collaborators Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. - PubMed
-
- Gudmundsson J, et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nat Genet. 2007;39:631–637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

