Yeast prions form infectious amyloid inclusion bodies in bacteria
- PMID: 22731490
- PMCID: PMC3520751
- DOI: 10.1186/1475-2859-11-89
Yeast prions form infectious amyloid inclusion bodies in bacteria
Abstract
Background: Prions were first identified as infectious proteins associated with fatal brain diseases in mammals. However, fungal prions behave as epigenetic regulators that can alter a range of cellular processes. These proteins propagate as self-perpetuating amyloid aggregates being an example of structural inheritance. The best-characterized examples are the Sup35 and Ure2 yeast proteins, corresponding to [PSI+] and [URE3] phenotypes, respectively.
Results: Here we show that both the prion domain of Sup35 (Sup35-NM) and the Ure2 protein (Ure2p) form inclusion bodies (IBs) displaying amyloid-like properties when expressed in bacteria. These intracellular aggregates template the conformational change and promote the aggregation of homologous, but not heterologous, soluble prionogenic molecules. Moreover, in the case of Sup35-NM, purified IBs are able to induce different [PSI+] phenotypes in yeast, indicating that at least a fraction of the protein embedded in these deposits adopts an infectious prion fold.
Conclusions: An important feature of prion inheritance is the existence of strains, which are phenotypic variants encoded by different conformations of the same polypeptide. We show here that the proportion of infected yeast cells displaying strong and weak [PSI+] phenotypes depends on the conditions under which the prionogenic aggregates are formed in E. coli, suggesting that bacterial systems might become useful tools to generate prion strain diversity.
Figures


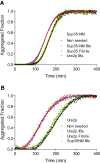




Similar articles
-
A promiscuous prion: efficient induction of [URE3] prion formation by heterologous prion domains.Genetics. 2009 Nov;183(3):929-40. doi: 10.1534/genetics.109.109322. Epub 2009 Sep 14. Genetics. 2009. PMID: 19752212 Free PMC article.
-
Human J-protein DnaJB6b Cures a Subset of Saccharomyces cerevisiae Prions and Selectively Blocks Assembly of Structurally Related Amyloids.J Biol Chem. 2016 Feb 19;291(8):4035-47. doi: 10.1074/jbc.M115.700393. Epub 2015 Dec 23. J Biol Chem. 2016. PMID: 26702057 Free PMC article.
-
Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro.Proc Natl Acad Sci U S A. 2004 Aug 31;101(35):12934-9. doi: 10.1073/pnas.0404968101. Epub 2004 Aug 23. Proc Natl Acad Sci U S A. 2004. PMID: 15326312 Free PMC article.
-
Prions are affected by evolution at two levels.Cell Mol Life Sci. 2016 Mar;73(6):1131-44. doi: 10.1007/s00018-015-2109-6. Epub 2015 Dec 28. Cell Mol Life Sci. 2016. PMID: 26713322 Free PMC article. Review.
-
A bipolar personality of yeast prion proteins.Prion. 2011 Oct-Dec;5(4):305-10. doi: 10.4161/pri.18307. Epub 2011 Oct 1. Prion. 2011. PMID: 22156730 Free PMC article. Review.
Cited by
-
Why and how protein aggregation has to be studied in vivo.Microb Cell Fact. 2013 Feb 15;12:17. doi: 10.1186/1475-2859-12-17. Microb Cell Fact. 2013. PMID: 23410248 Free PMC article.
-
Engineered bacterial hydrophobic oligopeptide repeats in a synthetic yeast prion, [REP-PSI (+)].Front Microbiol. 2015 Apr 21;6:311. doi: 10.3389/fmicb.2015.00311. eCollection 2015. Front Microbiol. 2015. PMID: 25954252 Free PMC article.
-
The prion-like RNA-processing protein HNRPDL forms inherently toxic amyloid-like inclusion bodies in bacteria.Microb Cell Fact. 2015 Jul 11;14:102. doi: 10.1186/s12934-015-0284-7. Microb Cell Fact. 2015. PMID: 26160665 Free PMC article.
-
Prion propagation can occur in a prokaryote and requires the ClpB chaperone.Elife. 2014 Aug 13;3:e02949. doi: 10.7554/eLife.02949. Elife. 2014. PMID: 25122461 Free PMC article.
-
Computational analysis of candidate prion-like proteins in bacteria and their role.Front Microbiol. 2015 Oct 15;6:1123. doi: 10.3389/fmicb.2015.01123. eCollection 2015. Front Microbiol. 2015. PMID: 26528269 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

