In silico analysis of HLA associations with drug-induced liver injury: use of a HLA-genotyped DNA archive from healthy volunteers
- PMID: 22732016
- PMCID: PMC3698530
- DOI: 10.1186/gm350
In silico analysis of HLA associations with drug-induced liver injury: use of a HLA-genotyped DNA archive from healthy volunteers
Abstract
Background: Drug-induced liver injury (DILI) is one of the most common adverse reactions leading to product withdrawal post-marketing. Recently, genome-wide association studies have identified a number of human leukocyte antigen (HLA) alleles associated with DILI; however, the cellular and chemical mechanisms are not fully understood.
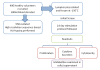
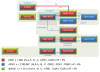
Methods: To study these mechanisms, we established an HLA-typed cell archive from 400 healthy volunteers. In addition, we utilized HLA genotype data from more than four million individuals from publicly accessible repositories such as the Allele Frequency Net Database, Major Histocompatibility Complex Database and Immune Epitope Database to study the HLA alleles associated with DILI. We utilized novel in silico strategies to examine HLA haplotype relationships among the alleles associated with DILI by using bioinformatics tools such as NetMHCpan, PyPop, GraphViz, PHYLIP and TreeView.
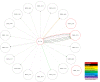
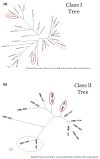
Results: We demonstrated that many of the alleles that have been associated with liver injury induced by structurally diverse drugs (flucloxacillin, co-amoxiclav, ximelagatran, lapatinib, lumiracoxib) reside on common HLA haplotypes, which were present in populations of diverse ethnicity.
Conclusions: Our bioinformatic analysis indicates that there may be a connection between the different HLA alleles associated with DILI caused by therapeutically and structurally different drugs, possibly through peptide binding of one of the HLA alleles that defines the causal haplotype. Further functional work, together with next-generation sequencing techniques, will be needed to define the causal alleles associated with DILI.
Figures
Similar articles
-
Impact of SLCO1B1*5 on Flucloxacillin and Co-Amoxiclav-Related Liver Injury.Front Pharmacol. 2022 Jun 8;13:882962. doi: 10.3389/fphar.2022.882962. eCollection 2022. Front Pharmacol. 2022. PMID: 35754504 Free PMC article.
-
Drug-induced liver injury: past, present and future.Pharmacogenomics. 2010 May;11(5):607-11. doi: 10.2217/pgs.10.24. Pharmacogenomics. 2010. PMID: 20415545 Review.
-
Minocycline hepatotoxicity: Clinical characterization and identification of HLA-B∗35:02 as a risk factor.J Hepatol. 2017 Jul;67(1):137-144. doi: 10.1016/j.jhep.2017.03.010. Epub 2017 Mar 18. J Hepatol. 2017. PMID: 28323125 Free PMC article.
-
Human leukocyte antigen genetic risk factors of drug-induced liver toxicology.Expert Opin Drug Metab Toxicol. 2015 Mar;11(3):395-409. doi: 10.1517/17425255.2015.992414. Epub 2014 Dec 10. Expert Opin Drug Metab Toxicol. 2015. PMID: 25491399 Review.
-
HLA-DR7 and HLA-DQ2: Transgenic mouse strains tested as a model system for ximelagatran hepatotoxicity.PLoS One. 2017 Sep 21;12(9):e0184744. doi: 10.1371/journal.pone.0184744. eCollection 2017. PLoS One. 2017. PMID: 28934241 Free PMC article.
Cited by
-
Case Characterization, Clinical Features and Risk Factors in Drug-Induced Liver Injury.Int J Mol Sci. 2016 May 12;17(5):714. doi: 10.3390/ijms17050714. Int J Mol Sci. 2016. PMID: 27187363 Free PMC article. Review.
-
Selective HLA Class II Allele-Restricted Activation of Atabecestat Metabolite-Specific Human T-Cells.Chem Res Toxicol. 2024 Oct 21;37(10):1712-1727. doi: 10.1021/acs.chemrestox.4c00262. Epub 2024 Sep 30. Chem Res Toxicol. 2024. PMID: 39348529 Free PMC article.
-
Genetics of immune-mediated adverse drug reactions: a comprehensive and clinical review.Clin Rev Allergy Immunol. 2015 Jun;48(2-3):165-75. doi: 10.1007/s12016-014-8418-y. Clin Rev Allergy Immunol. 2015. PMID: 24777842 Review.
-
HLA class I and class II conserved extended haplotypes and their fragments or blocks in Mexicans: implications for the study of genetic diversity in admixed populations.PLoS One. 2013 Sep 23;8(9):e74442. doi: 10.1371/journal.pone.0074442. eCollection 2013. PLoS One. 2013. PMID: 24086347 Free PMC article.
-
Identification of 8-Digit HLA-A, -B, -C, and -DRB1 Allele and Haplotype Frequencies in Koreans Using the One Lambda AllType Next-Generation Sequencing Kit.Ann Lab Med. 2021 May 1;41(3):310-317. doi: 10.3343/alm.2021.41.3.310. Ann Lab Med. 2021. PMID: 33303716 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

