Molecular chaperones DnaK and DnaJ share predicted binding sites on most proteins in the E. coli proteome
- PMID: 22732719
- PMCID: PMC3462289
- DOI: 10.1039/c2mb25145k
Molecular chaperones DnaK and DnaJ share predicted binding sites on most proteins in the E. coli proteome
Abstract
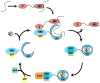
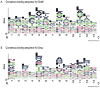
In Escherichia coli, the molecular chaperones DnaK and DnaJ cooperate to assist the folding of newly synthesized or unfolded polypeptides. DnaK and DnaJ bind to hydrophobic motifs in these proteins and they also bind to each other. Together, this system is thought to be sufficiently versatile to act on the entire proteome, which creates interesting challenges in understanding the interactions between DnaK, DnaJ and their thousands of potential substrates. To address this question, we computationally predicted the number and frequency of DnaK- and DnaJ-binding motifs in the E. coli proteome, guided by free energy-based binding consensus motifs. This analysis revealed that nearly every protein is predicted to contain multiple DnaK- and DnaJ-binding sites, with the DnaJ sites occurring approximately twice as often. Further, we found that an overwhelming majority of the DnaK sites partially or completely overlapped with the DnaJ-binding motifs. It is well known that high concentrations of DnaJ inhibit DnaK-DnaJ-mediated refolding. The observed overlapping binding sites suggest that this phenomenon may be explained by an important balance in the relative stoichiometry of DnaK and DnaJ. To test this idea, we measured the chaperone-assisted folding of two denatured substrates and found that the distribution of predicted DnaK- and DnaJ-binding sites was indeed a good predictor of the optimal stoichiometry required for folding. These studies provide insight into how DnaK and DnaJ might cooperate to maintain global protein homeostasis.
Figures


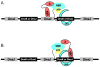


Similar articles
-
Molecular basis for regulation of the heat shock transcription factor sigma32 by the DnaK and DnaJ chaperones.Mol Cell. 2008 Nov 7;32(3):347-58. doi: 10.1016/j.molcel.2008.09.016. Mol Cell. 2008. PMID: 18995833
-
Recognizability of heterologous co-chaperones with Streptococcus intermedius DnaK and Escherichia coli DnaK.Microbiol Immunol. 2018 Nov;62(11):681-693. doi: 10.1111/1348-0421.12651. Epub 2018 Nov 5. Microbiol Immunol. 2018. PMID: 30239035
-
Regulation of ATPase and chaperone cycle of DnaK from Thermus thermophilus by the nucleotide exchange factor GrpE.J Mol Biol. 2001 Feb 2;305(5):1173-83. doi: 10.1006/jmbi.2000.4373. J Mol Biol. 2001. PMID: 11162122
-
Interferon-gamma is a target for binding and folding by both Escherichia coli chaperone model systems GroEL/GroES and DnaK/DnaJ/GrpE.Biochimie. 1998 Aug-Sep;80(8-9):729-37. doi: 10.1016/s0300-9084(99)80026-1. Biochimie. 1998. PMID: 9865495 Review.
-
The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones.Cell Mol Life Sci. 2006 Nov;63(22):2560-70. doi: 10.1007/s00018-006-6192-6. Cell Mol Life Sci. 2006. PMID: 16952052 Free PMC article. Review.
Cited by
-
The Impact of Hidden Structure on Aggregate Disassembly by Molecular Chaperones.Front Mol Biosci. 2022 Jul 7;9:915307. doi: 10.3389/fmolb.2022.915307. eCollection 2022. Front Mol Biosci. 2022. PMID: 35874607 Free PMC article.
-
Computationally-Aided Modeling of Hsp70-Client Interactions: Past, Present, and Future.J Phys Chem B. 2022 Sep 15;126(36):6780-6791. doi: 10.1021/acs.jpcb.2c03806. Epub 2022 Aug 30. J Phys Chem B. 2022. PMID: 36040440 Free PMC article.
-
Heat Shock Protein DnaJ in Pseudomonas aeruginosa Affects Biofilm Formation via Pyocyanin Production.Microorganisms. 2020 Mar 12;8(3):395. doi: 10.3390/microorganisms8030395. Microorganisms. 2020. PMID: 32178243 Free PMC article.
-
Infection-relevant conditions dictate differential versus coordinate expression of Salmonella chaperones and cochaperones.mBio. 2025 May 14;16(5):e0022725. doi: 10.1128/mbio.00227-25. Epub 2025 Mar 31. mBio. 2025. PMID: 40162747 Free PMC article.
-
Antimicrobial Effects of Free Nitrous Acid on Desulfovibrio vulgaris: Implications for Sulfide-Induced Corrosion of Concrete.Appl Environ Microbiol. 2016 Aug 30;82(18):5563-75. doi: 10.1128/AEM.01655-16. Print 2016 Sep 15. Appl Environ Microbiol. 2016. PMID: 27371588 Free PMC article.

