Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3
- PMID: 22732842
- PMCID: PMC3410389
- DOI: 10.1038/embor.2012.89
Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3
Abstract
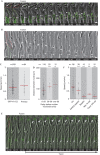
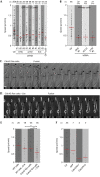
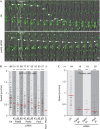
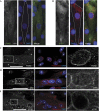
Cells actively position their nucleus within the cytoplasm. One striking example is observed during skeletal myogenesis. Differentiated myoblasts fuse to form a multinucleated myotube with nuclei positioned in the centre of the syncytium by an unknown mechanism. Here, we describe that the nucleus of a myoblast moves rapidly after fusion towards the central myotube nuclei. This movement is driven by microtubules and dynein/dynactin complex, and requires Cdc42, Par6 and Par3. We found that Par6β and dynactin accumulate at the nuclear envelope of differentiated myoblasts and myotubes, and this accumulation is dependent on Par6 and Par3 proteins but not on microtubules. These results suggest a mechanism where nuclear movement after fusion is driven by microtubules that emanate from one nucleus that are pulled by dynein/dynactin complex anchored to the nuclear envelope of another nucleus.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- Dauer WT, Worman HJ (2009) The Nuclear envelope as a signaling node in development and disease. Dev Cell 17: 626–638 - PubMed
-
- Reinsch S, Gönczy P (1998) Mechanisms of nuclear positioning. J Cell Sci 111: 2283–2295 - PubMed
-
- Gomes ER, Jani S, Gundersen GG (2005) Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell 121: 451–463 - PubMed
-
- Cappello S et al. (2006) The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat Neurosci 9: 1099–1107 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

