Genetic variations in T-cell activation and effector pathways modulate alloimmune responses after allogeneic hematopoietic stem cell transplantation in patients with hematologic malignancies
- PMID: 22733023
- PMCID: PMC3590086
- DOI: 10.3324/haematol.2012.066159
Genetic variations in T-cell activation and effector pathways modulate alloimmune responses after allogeneic hematopoietic stem cell transplantation in patients with hematologic malignancies
Abstract
Background: Recently, several important polymorphisms have been identified in T-cell activation and effector pathway genes and have been reported to be associated with inter-patient variability in alloimmune responses. The present study was designed to assess the impact of these genetic variations on the outcomes of allogeneic hematopoietic stem cell transplantation.
Design and methods: We first investigated ten single nucleotide polymorphisms in six genes, CD28, inducible co-stimulator, cytotoxic T-lymphocyte antigen 4, granzyme B, Fas and Fas ligand, in 138 pairs of patients and their unrelated donors and a second cohort of 102 pairs of patients and their HLA-identical sibling donors.
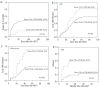
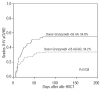
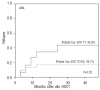
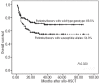
Results: We observed that patients receiving stem cells from a donor with the cytotoxic T-lymphocyte antigen 4 gene CT60 variant allele (AA genotype) had a reduced incidence of grades II-IV acute graft-versus-host disease; however, they experienced early cytomegalovirus infection and relapsed more frequently, which suggested an interaction between the donor cytotoxic T-lymphocyte antigen 4 gene CT60 AA genotype and reduced T-cell alloreactivity. Furthermore, an unrelated donor with the granzyme B +55 variant genotype (AA) was an independent risk factor for development of grades II-IV acute graft-versus-host disease (P=0.024, RR=1.811). Among patients with acute myelogenous leukemia, those with the Fas -670 TT genotype were at higher risk of relapse (P=0.003, RR=3.823). The presence of these susceptible alleles in the donor and/or patient resulted in worse overall survival (54.9% versus 69.5%, P=0.029).
Conclusions: Our data suggest that genotype analysis of T-cell activation and effector pathway genes can be used for risk assessment for patients with hematologic malignancies before hematopoietic stem cell transplantation.
Figures
References
-
- Penack O, Holler E, van den Brink., MR Graft-versus-host disease: regulation by microbe-associated molecules and innate immune receptors. Blood. 2010;115(10):1865-72 - PubMed
-
- Rocha V, Porcher R, Fernandes JF, Filion A, Bittencourt H, Silva W, Jr, et al. Association of drug metabolism gene polymorphisms with toxicities, graft-versus-host disease and survival after HLA-identical sibling hematopoietic stem cell transplantation for patients with leukemia. Leukemia. 2009;23(3):545-56 - PubMed
-
- Mayor NP, Shaw BE, Hughes DA, Maldonado-Torres H, Madrigal JA, Keshav S, et al. Single nucleotide polymorphisms in the NOD2/CARD15 gene are associated with an increased risk of relapse and death for patients with acute leukemia after hematopoietic stem-cell transplantation with unrelated donors. J Clin Oncol. 2007;25(27):4262-9 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

