Role of ISKpn7 and deletions in blaKPC gene expression
- PMID: 22733068
- PMCID: PMC3421896
- DOI: 10.1128/AAC.00334-12
Role of ISKpn7 and deletions in blaKPC gene expression
Abstract
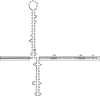
The carbapenemase-encoding bla(KPC) gene, which is rapidly spreading in Gram-negative rods, is located on a Tn3-based transposon, Tn4401, which carries a polymorphic region giving rise to five isoforms (a, b, c, d, and e) that is located immediately upstream of the bla(KPC) gene and thus likely involved in its expression. Using 5' rapid amplification of cDNA ends (5'RACE), we identified three potential promoter sequences (P1, P2, and P3) upstream of the bla(KPC) gene, of which only P1 (absent from isoforms c and d) and P2 (present in all isoforms, with a -35 box located inside the right inverted repeat of ISKpn7) were shown to be true promoters involved in expression. One representative of each different promoter combination of Tn4401, i.e., P2 alone (isoform c), P1-P2 (isoform a), and P1-P2-P3 (isoform b), was cloned into an Escherichia coli plasmid vector. Using reverse transcription-PCR (RT-PCR), the highest level of expression was obtained with isoform a (P1 and P2), which is also the most commonly encountered form in enterobacterial clinical isolates, followed by isoforms b (P1, P2, and P3) and c (P2 only). These differences in expression led to slight differences in MIC values of carbapenems. In silico analysis of the DNA sequence of isoform b revealed a stem-loop structure that is likely responsible for strong stops observed in 5'RACE experiments and for decreased expression compared to that with isoform a (P1 and P2). In addition, such structures could also be at the origin for the deletions observed in isoforms a and c. Taken together, these results indicate that the P1 and P2 promoters both contribute to the expression of the bla(KPC) gene and that the construct with the highest level of expression is that possessing isoform a, which is also the most commonly encountered form in clinical isolates.
Figures


References
-
- Bradford PA, et al. 2004. Emergence of carbapenem-resistant Klebsiella species possessing the class A carbapenem-hydrolyzing KPC-2 and inhibitor-resistant TEM-30 β-lactamases in New York City. Clin. Infect. Dis. 39:55–60 - PubMed
-
- Browning DF, Busby SJ. 2004. The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2:57–65 - PubMed
-
- Clinical and Laboratory Standards Institute 2012. Performance standards for antimicrobial susceptibility testing; 21st informational supplement. CLSI M100-S21. Clinical and Laboratory Standards Institute, Wayne, PA
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

