Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase
- PMID: 22733076
- PMCID: PMC3421890
- DOI: 10.1128/AAC.00957-12
Severe acute respiratory syndrome coronavirus replication inhibitor that interferes with the nucleic acid unwinding of the viral helicase
Abstract
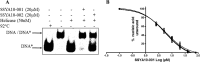
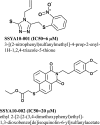
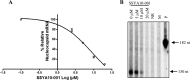
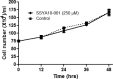
Severe acute respiratory syndrome (SARS) is a highly contagious disease, caused by SARS coronavirus (SARS-CoV), for which there are no approved treatments. We report the discovery of a potent inhibitor of SARS-CoV that blocks replication by inhibiting the unwinding activity of the SARS-CoV helicase (nsp13). We used a Förster resonance energy transfer (FRET)-based helicase assay to screen the Maybridge Hitfinder chemical library. We identified and validated a compound (SSYA10-001) that specifically blocks the double-stranded RNA (dsRNA) and dsDNA unwinding activities of nsp13, with 50% inhibitory concentrations (IC(50)s) of 5.70 and 5.30 μM, respectively. This compound also has inhibitory activity (50% effective concentration [EC(50)] = 8.95 μM) in a SARS-CoV replicon assay, with low cytotoxicity (50% cytotoxic concentration [CC(50)] = >250 μM), suggesting that the helicase plays a still unidentified critical role in the SARS-CoV life cycle. Enzyme kinetic studies on the mechanism of nsp13 inhibition revealed that SSYA10-001 acts as a noncompetitive inhibitor of nsp13 with respect to nucleic acid and ATP substrates. Moreover, SSYA10-001 does not affect ATP hydrolysis or nsp13 binding to the nucleic acid substrate. SSYA10-001 did not inhibit hepatitis C virus (HCV) helicase, other bacterial and viral RNA-dependent RNA polymerases, or reverse transcriptase. These results suggest that SSYA10-001 specifically blocks nsp13 through a novel mechanism and is less likely to interfere with the functions of cellular enzymes that process nucleic acids or ATP. Hence, it is possible that SSYA10-001 inhibits unwinding by nsp13 by affecting conformational changes during the course of the reaction or translocation on the nucleic acid. SSYA10-001 will be a valuable tool for studying the specific role of nsp13 in the SARS-CoV life cycle, which could be a model for other nidoviruses and also a candidate for further development as a SARS antiviral target.
Figures




References
-
- Abdel-Megeed AM, Abdel-Rahman HM, Alkaramany GE, El-Gendy MA. 2009. Design, synthesis and molecular modeling study of acylated 1,2,4-triazole-3-acetates with potential anti-inflammatory activity. Eur. J. Med. Chem. 44:117–123 - PubMed
-
- Ahnert P, Patel SS. 1997. Asymmetric interactions of hexameric bacteriophage T7 DNA helicase with the 5′- and 3′-tails of the forked DNA substrate. J. Biol. Chem. 272:32267–32273 - PubMed
-
- Ali JA, Lohman TM. 1997. Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275:377–380 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

