Ume6 transcription factor is part of a signaling cascade that regulates autophagy
- PMID: 22733735
- PMCID: PMC3396506
- DOI: 10.1073/pnas.1200313109
Ume6 transcription factor is part of a signaling cascade that regulates autophagy
Abstract
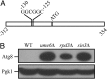
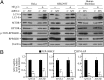
Autophagy has been implicated in a number of physiological processes important for human heath and disease. Autophagy involves the formation of a double-membrane cytosolic vesicle, an autophagosome. Central to the formation of the autophagosome is the ubiquitin-like protein autophagy-related (Atg)8 (microtubule-associated protein 1 light chain 3/LC3 in mammalian cells). Following autophagy induction, Atg8 shows the greatest change in expression of any of the proteins required for autophagy. The magnitude of autophagy is, in part, controlled by the amount of Atg8; thus, controlling Atg8 protein levels is one potential mechanism for modulating autophagy activity. We have identified a negative regulator of ATG8 transcription, Ume6, which acts along with a histone deacetylase complex including Sin3 and Rpd3 to regulate Atg8 levels; deletion of any of these components leads to an increase in Atg8 and a concomitant increase in autophagic activity. A similar regulatory mechanism is present in mammalian cells, indicating that this process is highly conserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
The Ume6-Sin3-Rpd3 complex regulates ATG8 transcription to control autophagosome size.Autophagy. 2012 Dec;8(12):1835-6. doi: 10.4161/auto.21845. Epub 2012 Sep 7. Autophagy. 2012. PMID: 22960621 Free PMC article.
-
The autophagy-related protein kinase Atg1 interacts with the ubiquitin-like protein Atg8 via the Atg8 family interacting motif to facilitate autophagosome formation.J Biol Chem. 2012 Aug 17;287(34):28503-7. doi: 10.1074/jbc.C112.387514. Epub 2012 Jul 9. J Biol Chem. 2012. PMID: 22778255 Free PMC article.
-
The trehalose-6-phosphate phosphatase Tps2 regulates ATG8 transcription and autophagy in Saccharomyces cerevisiae.Autophagy. 2021 Apr;17(4):1013-1027. doi: 10.1080/15548627.2020.1746592. Epub 2020 Apr 2. Autophagy. 2021. PMID: 32240040 Free PMC article.
-
Role of the mammalian ATG8/LC3 family in autophagy: differential and compensatory roles in the spatiotemporal regulation of autophagy.BMB Rep. 2016 Aug;49(8):424-30. doi: 10.5483/bmbrep.2016.49.8.081. BMB Rep. 2016. PMID: 27418283 Free PMC article. Review.
-
Transcriptional regulation of ATG9 by the Pho23-Rpd3 complex modulates the frequency of autophagosome formation.Autophagy. 2014 Sep;10(9):1681-2. doi: 10.4161/auto.29641. Epub 2014 Jul 7. Autophagy. 2014. PMID: 25046109 Free PMC article. Review.
Cited by
-
The RNA polymerase II subunit Rpb9 activates ATG1 transcription and autophagy.EMBO Rep. 2022 Nov 7;23(11):e54993. doi: 10.15252/embr.202254993. Epub 2022 Sep 14. EMBO Rep. 2022. PMID: 36102592 Free PMC article.
-
Cdk8 Kinase Module: A Mediator of Life and Death Decisions in Times of Stress.Microorganisms. 2021 Oct 15;9(10):2152. doi: 10.3390/microorganisms9102152. Microorganisms. 2021. PMID: 34683473 Free PMC article. Review.
-
Atg8 family proteins, LIR/AIM motifs and other interaction modes.Autophagy Rep. 2023 Mar 19;2(1):2188523. doi: 10.1080/27694127.2023.2188523. eCollection 2023 Dec 31. Autophagy Rep. 2023. PMID: 38214012 Free PMC article.
-
Snf1 AMPK positively regulates ER-phagy via expression control of Atg39 autophagy receptor in yeast ER stress response.PLoS Genet. 2020 Sep 28;16(9):e1009053. doi: 10.1371/journal.pgen.1009053. eCollection 2020 Sep. PLoS Genet. 2020. PMID: 32986716 Free PMC article.
-
Distinct H3K27me3 and H3K27ac Modifications in Neural Tube Defects Induced by Benzo[a]pyrene.Brain Sci. 2023 Feb 15;13(2):334. doi: 10.3390/brainsci13020334. Brain Sci. 2023. PMID: 36831877 Free PMC article.
References
-
- Xie Z, Klionsky DJ. Autophagosome formation: Core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

