Evolutionarily conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis
- PMID: 22733738
- PMCID: PMC3396513
- DOI: 10.1073/pnas.1119173109
Evolutionarily conserved glycan signal to degrade aberrant brassinosteroid receptors in Arabidopsis
Abstract
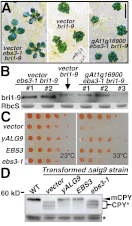
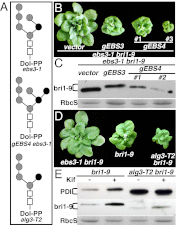
Asparagine-linked glycans (N-glycans) are crucial signals for protein folding, quality control, and endoplasmic reticulum (ER)-associated degradation (ERAD) in yeast and mammals. Although similar ERAD processes were reported in plants, little is known about their biochemical mechanisms, especially their relationships with N-glycans. Here, we show that a missense mutation in the Arabidopsis EMS-mutagenized bri1 suppressor 3 (EBS3) gene suppresses a dwarf mutant, bri1-9, the phenotypes of which are caused by ER retention and ERAD of a brassinosteroid receptor, BRASSINOSTEROID-INSENSITIVE 1 (BR1). EBS3 encodes the Arabidopsis ortholog of the yeast asparagine-linked glycosylation 9 (ALG9), which catalyzes the ER luminal addition of two terminal α1,2 mannose (Man) residues in assembling the three-branched N-glycan precursor [glucose(Glc)](3)(Man)(9)[N-acetylglucosamine(GlcNAc)](2). Consistent with recent discoveries revealing the importance of the Glc(3)Man(9)GlcNAc(2) C-branch in generating an ERAD signal, the ebs3-1 mutation prevents the Glc(3)Man(9)GlcNAc(2) assembly and inhibits the ERAD of bri1-9. By contrast, overexpression of EBS4 in ebs3-1 bri1-9, which encodes the Arabidopsis ortholog of the yeast ALG12 catalyzing the ER luminal α1,6 Man addition, adds an α1,6 Man to the truncated N-glycan precursor accumulated in ebs3-1 bri1-9, promotes the bri1-9 ERAD, and neutralizes the ebs3-1 suppressor phenotype. Furthermore, a transfer (T)-DNA insertional alg3-T2 mutation, which causes accumulation of an even smaller N-glycan precursor carrying a different exposed α1,6 Man, promotes the ERAD of bri1-9 and enhances its dwarfism. Taken together, our results strongly suggest that the glycan signal to mark an ERAD client in Arabidopsis is likely conserved to be an α1,6 Man-exposed N-glycan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Abu-Qarn M, Eichler J, Sharon N. Not just for Eukarya anymore: Protein glycosylation in Bacteria and Archaea. Curr Opin Struct Biol. 2008;18:544–550. - PubMed
-
- Kelleher DJ, Gilmore R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology. 2006;16:47R–62R. - PubMed
-
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. - PubMed
-
- Molinari M. N-glycan structure dictates extension of protein folding or onset of disposal. Nat Chem Biol. 2007;3:313–320. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

