Deamidation accelerates amyloid formation and alters amylin fiber structure
- PMID: 22734583
- PMCID: PMC3410046
- DOI: 10.1021/ja3039486
Deamidation accelerates amyloid formation and alters amylin fiber structure
Abstract
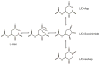
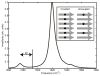
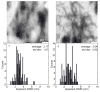
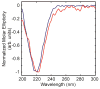
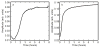
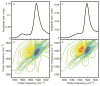
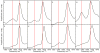
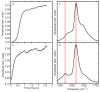
Deamidation of asparagine and glutamine is the most common nonenzymatic, post-translational modification. Deamidation can influence the structure, stability, folding, and aggregation of proteins and has been proposed to play a role in amyloid formation. However there are no structural studies of the consequences of deamidation on amyloid fibers, in large part because of the difficulty of studying these materials using conventional methods. Here we examine the effects of deamidation on the kinetics of amyloid formation by amylin, the causative agent of type 2 diabetes. We find that deamidation accelerates amyloid formation and the deamidated material is able to seed amyloid formation by unmodified amylin. Using site-specific isotope labeling and two-dimensional infrared (2D IR) spectroscopy, we show that fibers formed by samples that contain deamidated polypeptide contain reduced amounts of β-sheet. Deamidation leads to disruption of the N-terminal β-sheet between Ala-8 and Ala-13, but β-sheet is still retained near Leu-16. The C-terminal sheet is disrupted near Leu-27. Analysis of potential sites of deamidation together with structural models of amylin fibers reveals that deamidation in the N-terminal β-sheet region may be the cause for the disruption of the fiber structure at both the N- and C-terminal β-sheet. Thus, deamidation is a post-translational modification that creates fibers that have an altered structure but can still act as a template for amylin aggregation. Deamidation is very difficult to detect with standard methods used to follow amyloid formation, but isotope-labeled IR spectroscopy provides a means for monitoring sample degradation and investigating the structural consequences of deamidation.
Figures

Similar articles
-
Low levels of asparagine deamidation can have a dramatic effect on aggregation of amyloidogenic peptides: implications for the study of amyloid formation.Protein Sci. 2002 Feb;11(2):342-9. doi: 10.1110/ps.48702. Protein Sci. 2002. PMID: 11790844 Free PMC article.
-
Two-dimensional infrared spectroscopy reveals the complex behaviour of an amyloid fibril inhibitor.Nat Chem. 2012 Mar 11;4(5):355-60. doi: 10.1038/nchem.1293. Nat Chem. 2012. PMID: 22522254 Free PMC article.
-
Structural polymorphism of human islet amyloid polypeptide (hIAPP) oligomers highlights the importance of interfacial residue interactions.Biomacromolecules. 2011 Jan 10;12(1):210-20. doi: 10.1021/bm101159p. Epub 2010 Dec 15. Biomacromolecules. 2011. PMID: 21158384
-
Islet amyloid: from fundamental biophysics to mechanisms of cytotoxicity.FEBS Lett. 2013 Apr 17;587(8):1106-18. doi: 10.1016/j.febslet.2013.01.046. Epub 2013 Feb 1. FEBS Lett. 2013. PMID: 23380070 Free PMC article. Review.
-
Causative factors for formation of toxic islet amyloid polypeptide oligomer in type 2 diabetes mellitus.Clin Interv Aging. 2015 Nov 19;10:1873-9. doi: 10.2147/CIA.S95297. eCollection 2015. Clin Interv Aging. 2015. PMID: 26604727 Free PMC article. Review.
Cited by
-
Aggregation of islet amyloid polypeptide: from physical chemistry to cell biology.Curr Opin Struct Biol. 2013 Feb;23(1):82-9. doi: 10.1016/j.sbi.2012.11.003. Epub 2012 Dec 22. Curr Opin Struct Biol. 2013. PMID: 23266002 Free PMC article. Review.
-
Implications of Metal Binding and Asparagine Deamidation for Amyloid Formation.Int J Mol Sci. 2018 Aug 19;19(8):2449. doi: 10.3390/ijms19082449. Int J Mol Sci. 2018. PMID: 30126231 Free PMC article. Review.
-
Transparent window 2D IR spectroscopy of proteins.J Chem Phys. 2021 Jul 28;155(4):040903. doi: 10.1063/5.0052628. J Chem Phys. 2021. PMID: 34340394 Free PMC article. Review.
-
Islet Amyloid Polypeptide: Structure, Function, and Pathophysiology.J Diabetes Res. 2016;2016:2798269. doi: 10.1155/2016/2798269. Epub 2015 Nov 15. J Diabetes Res. 2016. PMID: 26649319 Free PMC article. Review.
-
Heat-induced irreversible denaturation of the camelid single domain VHH antibody is governed by chemical modifications.J Biol Chem. 2014 May 30;289(22):15666-79. doi: 10.1074/jbc.M113.534222. Epub 2014 Apr 16. J Biol Chem. 2014. PMID: 24739391 Free PMC article.
References
-
- Walsh CT, Garneau-Tsodikova S, Gatto GJ. Angew Chem Int Ed. 2005;44:7342–7372. - PubMed
-
- Mann M, Jensen ON. Nat Biotechnol. 2003;21:255–261. - PubMed
-
- Witze ES, Old WM, Resing KA, Ahn NG. Nat Methods. 2007;4:798–06. - PubMed
-
- Bischoff R, Kolbe HVJ. J Chromatogr B: Biomed Sci App. 1994;662:261–78. - PubMed
-
- Geiger T, Clarke S. J Biol Chem. 1987;262:785–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

