Cotranslational protein folding within the ribosome tunnel influences trigger-factor recruitment
- PMID: 22735532
- PMCID: PMC3379017
- DOI: 10.1016/j.bpj.2012.04.048
Cotranslational protein folding within the ribosome tunnel influences trigger-factor recruitment
Abstract
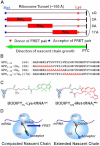
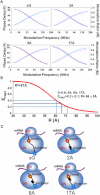
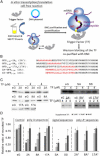
In recent years, various folding zones within the ribosome tunnel have been identified and explored through x-ray, cryo-electron microscopy (cryo-EM), and molecular biology studies. Here, we generated ribosome-bound nascent polypeptide complexes (RNCs) with different polyalanine (poly-A) inserts or signal peptides from membrane/secretory proteins to explore the influence of nascent chain compaction in the Escherichia coli ribosome tunnel on chaperone recruitment. By employing time-resolved fluorescence resonance energy transfer and immunoblotting, we were able to show that the poly-A inserts embedded in the passage tunnel can form a compacted structure (presumably helix) and reduce the recruitment of Trigger Factor (TF) when the helical motif is located in the region near the tunnel exit. Similar experiments on nascent chains containing signal sequences that may form compacted structural motifs within the ribosome tunnel and lure the signal recognition particle (SRP) to the ribosome, provided additional evidence that short, compacted nascent chains interfere with TF binding. These findings shed light on the possible controlling mechanism of nascent chains within the tunnel that leads to chaperone recruitment, as well as the function of L23, the ribosomal protein that serves as docking sites for both TF and SRP, in cotranslational protein targeting.
Copyright © 2012 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Figures


Similar articles
-
Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome.J Cell Biol. 2003 May 26;161(4):679-84. doi: 10.1083/jcb.200302130. Epub 2003 May 19. J Cell Biol. 2003. PMID: 12756233 Free PMC article.
-
Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor.Proc Natl Acad Sci U S A. 2004 May 25;101(21):7902-6. doi: 10.1073/pnas.0402231101. Epub 2004 May 17. Proc Natl Acad Sci U S A. 2004. PMID: 15148364 Free PMC article.
-
Global profiling of SRP interaction with nascent polypeptides.Nature. 2016 Aug 11;536(7615):219-23. doi: 10.1038/nature19070. Epub 2016 Aug 3. Nature. 2016. PMID: 27487212
-
Cryo-electron microscopy of ribosomal complexes in cotranslational folding, targeting, and translocation.Wiley Interdiscip Rev RNA. 2012 May-Jun;3(3):429-41. doi: 10.1002/wrna.119. Epub 2011 Nov 17. Wiley Interdiscip Rev RNA. 2012. PMID: 22095783 Review.
-
Cotranslational processing mechanisms: towards a dynamic 3D model.Trends Biochem Sci. 2009 Aug;34(8):417-26. doi: 10.1016/j.tibs.2009.04.003. Epub 2009 Jul 31. Trends Biochem Sci. 2009. PMID: 19647435 Review.
Cited by
-
Nonorthogonal tRNA(cys)(Amber) for protein and nascent chain labeling.RNA. 2015 Sep;21(9):1672-82. doi: 10.1261/rna.051805.115. Epub 2015 Jul 20. RNA. 2015. PMID: 26194135 Free PMC article.
-
Cotranslational Protein Folding inside the Ribosome Exit Tunnel.Cell Rep. 2015 Sep 8;12(10):1533-40. doi: 10.1016/j.celrep.2015.07.065. Epub 2015 Aug 28. Cell Rep. 2015. PMID: 26321634 Free PMC article.
-
Exploring Allosteric Signaling in the Exit Tunnel of the Bacterial Ribosome by Molecular Dynamics Simulations and Residue Network Model.Front Mol Biosci. 2020 Sep 25;7:586075. doi: 10.3389/fmolb.2020.586075. eCollection 2020. Front Mol Biosci. 2020. PMID: 33102529 Free PMC article.
-
Mechanisms governing codon usage bias and the implications for protein expression in the chloroplast of Chlamydomonas reinhardtii.Plant J. 2022 Nov;112(4):919-945. doi: 10.1111/tpj.15970. Epub 2022 Oct 19. Plant J. 2022. PMID: 36071273 Free PMC article. Review.
-
Strong anion-exchange fast performance liquid chromatography as a versatile tool for preparation and purification of RNA produced by in vitro transcription.RNA. 2013 Oct;19(10):1449-59. doi: 10.1261/rna.038117.113. Epub 2013 Aug 8. RNA. 2013. PMID: 23929938 Free PMC article.
References
-
- Kramer G., Boehringer D., Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat. Struct. Mol. Biol. 2009;16:589–597. - PubMed
-
- Ban N., Nissen P., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. - PubMed
-
- Voss N.R., Gerstein M., Moore P.B. The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 2006;360:893–906. - PubMed
-
- Beckmann R., Spahn C.M., Blobel G. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell. 2001;107:361–372. - PubMed
-
- Ferbitz L., Maier T., Ban N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–596. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

