Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing
- PMID: 22735697
- PMCID: PMC3458544
- DOI: 10.1093/nar/gks590
Inverted Alu dsRNA structures do not affect localization but can alter translation efficiency of human mRNAs independent of RNA editing
Abstract
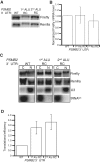
With over one million copies, Alu elements are the most abundant repetitive elements in the human genome. When transcribed, interaction between two Alus that are in opposite orientation gives rise to double-stranded RNA (dsRNA). Although the presence of dsRNA in the cell was previously thought to only occur during viral infection, it is now known that cells express many endogenous small dsRNAs, such as short interfering RNA (siRNAs) and microRNA (miRNAs), which regulate gene expression. It is possible that long dsRNA structures formed from Alu elements influence gene expression. Here, we report that human mRNAs containing inverted Alu elements are present in the mammalian cytoplasm. The presence of these long intramolecular dsRNA structures within 3'-UTRs decreases translational efficiency, and although the structures undergo extensive editing in vivo, the effects on translation are independent of the presence of inosine. As inverted Alus are predicted to reside in >5% of human protein-coding genes, these intramolecular dsRNA structures are important regulators of gene expression.
Figures



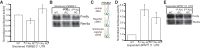
References
-
- Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161:529–540. - PubMed
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. - PubMed
-
- Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–607. - PubMed
-
- Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

