A polyglutamine expansion disease protein sequesters PTIP to attenuate DNA repair and increase genomic instability
- PMID: 22736030
- PMCID: PMC3441122
- DOI: 10.1093/hmg/dds246
A polyglutamine expansion disease protein sequesters PTIP to attenuate DNA repair and increase genomic instability
Abstract
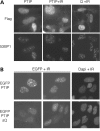
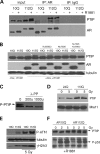
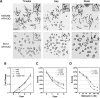
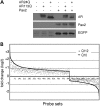
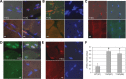
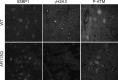
Glutamine (Q) expansion diseases are a family of degenerative disorders caused by the lengthening of CAG triplet repeats present in the coding sequences of seemingly unrelated genes whose mutant proteins drive pathogenesis. Despite all the molecular evidence for the genetic basis of these diseases, how mutant poly-Q proteins promote cell death and drive pathogenesis remains controversial. In this report, we show a specific interaction between the mutant androgen receptor (AR), a protein associated with spinal and bulbar muscular atrophy (SBMA), and the nuclear protein PTIP (Pax Transactivation-domain Interacting Protein), a protein with an unusually long Q-rich domain that functions in DNA repair. Upon exposure to ionizing radiation, PTIP localizes to nuclear foci that are sites of DNA damage and repair. However, the expression of poly-Q AR sequesters PTIP away from radiation-induced nuclear foci. This results in sensitivity to DNA-damaging agents and chromosomal instabilities. In a mouse model of SBMA, evidence for DNA damage is detected in muscle cell nuclei and muscular atrophy is accelerated when one copy of the gene encoding PTIP is removed. These data provide a new paradigm for understanding the mechanisms of cellular degeneration observed in poly-Q expansion diseases.
Figures



Similar articles
-
X-Linked Spinal and Bulbar Muscular Atrophy: From Clinical Genetic Features and Molecular Pathology to Mechanisms Underlying Disease Toxicity.Adv Exp Med Biol. 2018;1049:103-133. doi: 10.1007/978-3-319-71779-1_5. Adv Exp Med Biol. 2018. PMID: 29427100 Review.
-
In Vitro and In Vivo Modeling of Spinal and Bulbar Muscular Atrophy.J Mol Neurosci. 2016 Mar;58(3):365-73. doi: 10.1007/s12031-015-0677-4. Epub 2015 Nov 27. J Mol Neurosci. 2016. PMID: 26614347 Review.
-
Role of the androgen receptor CAG repeat polymorphism in prostate cancer, and spinal and bulbar muscular atrophy.Life Sci. 2011 Mar 28;88(13-14):565-71. doi: 10.1016/j.lfs.2011.01.021. Epub 2011 Jan 31. Life Sci. 2011. PMID: 21284948 Review.
-
Proteasomal and autophagic degradative activities in spinal and bulbar muscular atrophy.Neurobiol Dis. 2010 Nov;40(2):361-9. doi: 10.1016/j.nbd.2010.06.016. Epub 2010 Jul 16. Neurobiol Dis. 2010. PMID: 20621188 Review.
-
Molecular pathophysiology and disease-modifying therapies for spinal and bulbar muscular atrophy.Arch Neurol. 2012 Apr;69(4):436-40. doi: 10.1001/archneurol.2011.2308. Epub 2011 Dec 12. Arch Neurol. 2012. PMID: 22158719
Cited by
-
CAG RNAs induce DNA damage and apoptosis by silencing NUDT16 expression in polyglutamine degeneration.Proc Natl Acad Sci U S A. 2021 May 11;118(19):e2022940118. doi: 10.1073/pnas.2022940118. Proc Natl Acad Sci U S A. 2021. PMID: 33947817 Free PMC article.
-
Molecular Mechanisms and Therapeutics for SBMA/Kennedy's Disease.Neurotherapeutics. 2019 Oct;16(4):928-947. doi: 10.1007/s13311-019-00790-9. Neurotherapeutics. 2019. PMID: 31686397 Free PMC article. Review.
-
Molecular mechanisms underlying nucleotide repeat expansion disorders.Nat Rev Mol Cell Biol. 2021 Sep;22(9):589-607. doi: 10.1038/s41580-021-00382-6. Epub 2021 Jun 17. Nat Rev Mol Cell Biol. 2021. PMID: 34140671 Free PMC article. Review.
-
Determining the Fate of Neurons in SCA3: ATX3, a Rising Decision Maker in Response to DNA Stresses and Beyond.Front Cell Dev Biol. 2020 Dec 23;8:619911. doi: 10.3389/fcell.2020.619911. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33425926 Free PMC article. Review.
-
The central role of DNA damage and repair in CAG repeat diseases.Dis Model Mech. 2018 Jan 30;11(1):dmm031930. doi: 10.1242/dmm.031930. Dis Model Mech. 2018. PMID: 29419417 Free PMC article. Review.
References
-
- Takahashi T., Katada S., Onodera O. Polyglutamine diseases: where does toxicity come from? what is toxicity? where are we going? J. Mol. Cell. Biol. 2010;2:180–191. - PubMed
-
- Takeyama K., Ito S., Yamamoto A., Tanimoto H., Furutani T., Kanuka H., Miura M., Tabata T., Kato S. Androgen-dependent neurodegeneration by polyglutamine-expanded human androgen receptor in Drosophila. Neuron. 2002;35:855–864. - PubMed
-
- Klement I.A., Skinner P.J., Kaytor M.D., Yi H., Hersch S.M., Clark H.B., Zoghbi H.Y., Orr H.T. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95:41–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

