Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection
- PMID: 22737072
- PMCID: PMC3380936
- DOI: 10.1371/journal.ppat.1002769
Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection
Abstract
Intracellular pathogens such as Mycobacterium tuberculosis have evolved strategies for coping with the pressures encountered inside host cells. The ability to coordinate global gene expression in response to environmental and internal cues is one key to their success. Prolonged survival and replication within macrophages, a key virulence trait of M. tuberculosis, requires dynamic adaptation to diverse and changing conditions within its phagosomal niche. However, the physiological adaptations during the different phases of this infection process remain poorly understood. To address this knowledge gap, we have developed a multi-tiered approach to define the temporal patterns of gene expression in M. tuberculosis in a macrophage infection model that extends from infection, through intracellular adaptation, to the establishment of a productive infection. Using a clock plasmid to measure intracellular replication and death rates over a 14-day infection and electron microscopy to define bacterial integrity, we observed an initial period of rapid replication coupled with a high death rate. This was followed by period of slowed growth and enhanced intracellular survival, leading finally to an extended period of net growth. The transcriptional profiles of M. tuberculosis reflect these physiological transitions as the bacterium adapts to conditions within its host cell. Finally, analysis with a Transcriptional Regulatory Network model revealed linked genetic networks whereby M. tuberculosis coordinates global gene expression during intracellular survival. The integration of molecular and cellular biology together with transcriptional profiling and systems analysis offers unique insights into the host-driven responses of intracellular pathogens such as M. tuberculosis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
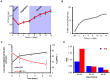
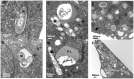
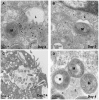
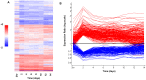
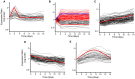
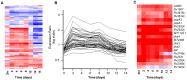

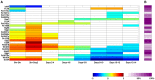
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

