The regulatory network of natural competence and transformation of Vibrio cholerae
- PMID: 22737089
- PMCID: PMC3380833
- DOI: 10.1371/journal.pgen.1002778
The regulatory network of natural competence and transformation of Vibrio cholerae
Abstract
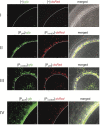
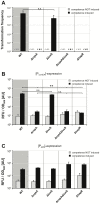
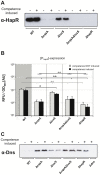
The human pathogen Vibrio cholerae is an aquatic bacterium frequently encountered in rivers, lakes, estuaries, and coastal regions. Within these environmental reservoirs, the bacterium is often found associated with zooplankton and more specifically with their chitinous exoskeleton. Upon growth on such chitinous surfaces, V. cholerae initiates a developmental program termed "natural competence for genetic transformation." Natural competence for transformation is a mode of horizontal gene transfer in bacteria and contributes to the maintenance and evolution of bacterial genomes. In this study, we investigated competence gene expression within this organism at the single cell level. We provide evidence that under homogeneous inducing conditions the majority of the cells express competence genes. A more heterogeneous expression pattern was observable on chitin surfaces. We hypothesize that this was the case due to the heterogeneity around the chitin surface, which might vary extensively with respect to chitin degradation products and autoinducers; these molecules contribute to competence induction based on carbon catabolite repression and quorum-sensing pathways, respectively. Therefore, we investigated the contribution of these two signaling pathways to natural competence in detail using natural transformation assays, transcriptional reporter fusions, quantitative RT-PCR, and immunological detection of protein levels using Western blot analysis. The results illustrate that all tested competence genes are dependent on the transformation regulator TfoX. Furthermore, intracellular cAMP levels play a major role in natural transformation. Finally, we demonstrate that only a minority of genes involved in natural transformation are regulated in a quorum-sensing-dependent manner and that these genes determine the fate of the surrounding DNA. We conclude with a model of the regulatory circuit of chitin-induced natural competence in V. cholerae.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Bertuzzo E, Mari L, Righetto L, Gatto M, Casagrandi R, et al. Prediction of the spatial evolution and effects of control measures for the unfolding Haiti cholera outbreak. Geophys Res Lett. 2011;38 doi:10.1029/2011GL046823.
-
- Tuite AR, Tien J, Eisenberg M, Earn DJ, Ma J, et al. Cholera Epidemic in Haiti, 2010: Using a Transmission Model to Explain Spatial Spread of Disease and Identify Optimal Control Interventions. Ann Intern Med. 2011;154:593–601. - PubMed
-
- Rinaldo A, Blokesch M, Bertuzzo E, Mari L, Righetto L, et al. A transmission model of the 2010 cholera epidemic in haiti. Ann Intern Med. 2011;155:403–404. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

