The microbiome of Brazilian mangrove sediments as revealed by metagenomics
- PMID: 22737213
- PMCID: PMC3380894
- DOI: 10.1371/journal.pone.0038600
The microbiome of Brazilian mangrove sediments as revealed by metagenomics
Abstract
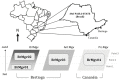
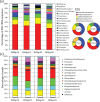
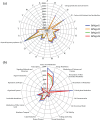
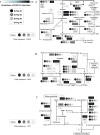
Here we embark in a deep metagenomic survey that revealed the taxonomic and potential metabolic pathways aspects of mangrove sediment microbiology. The extraction of DNA from sediment samples and the direct application of pyrosequencing resulted in approximately 215 Mb of data from four distinct mangrove areas (BrMgv01 to 04) in Brazil. The taxonomic approaches applied revealed the dominance of Deltaproteobacteria and Gammaproteobacteria in the samples. Paired statistical analysis showed higher proportions of specific taxonomic groups in each dataset. The metabolic reconstruction indicated the possible occurrence of processes modulated by the prevailing conditions found in mangrove sediments. In terms of carbon cycling, the sequences indicated the prevalence of genes involved in the metabolism of methane, formaldehyde, and carbon dioxide. With respect to the nitrogen cycle, evidence for sequences associated with dissimilatory reduction of nitrate, nitrogen immobilization, and denitrification was detected. Sequences related to the production of adenylsulfate, sulfite, and H(2)S were relevant to the sulphur cycle. These data indicate that the microbial core involved in methane, nitrogen, and sulphur metabolism consists mainly of Burkholderiaceae, Planctomycetaceae, Rhodobacteraceae, and Desulfobacteraceae. Comparison of our data to datasets from soil and sea samples resulted in the allotment of the mangrove sediments between those samples. The results of this study add valuable data about the composition of microbial communities in mangroves and also shed light on possible transformations promoted by microbial organisms in mangrove sediments.
Conflict of interest statement
Figures


References
-
- Sahoo K, Dhal N. Potencial microbial diversity in mangrove ecosystems: A review. IJMS 38(2) 2009;249:256.
-
- Duke NC, Meynecke J-O, Dittmann S, Ellison AM, Anger A, et al. A world without Mangroves? Science. 2007;317:41–42. - PubMed
-
- Holguin G, Vazquez P, Bashan Y. The role of sediment microorganisms in the productivity, conservation, and rehabilitation of mangrove ecosystems: an overview. Biol Fertil Soils. 2001;33:265–278.
-
- Ferreira TO, Otero XL, Souza-Junior VS, Vidal-Torrado P, Macías F, et al. Spatial patterns of soil attributes and components in a mangrove system in Southeast Brazil (São Paulo). J Soils Sed. 2010;10(6):995–1006.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

