Copy number analysis of complement C4A, C4B and C4A silencing mutation by real-time quantitative polymerase chain reaction
- PMID: 22737222
- PMCID: PMC3380926
- DOI: 10.1371/journal.pone.0038813
Copy number analysis of complement C4A, C4B and C4A silencing mutation by real-time quantitative polymerase chain reaction
Erratum in
- PLoS One. 2012;7(9). doi:10.1371/annotation/d19ba035-dbf0-4e58-932a-52efaa8137f3
Abstract
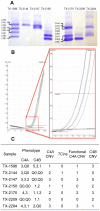
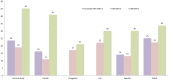
Low protein levels and copy number variation (CNV) of the fourth component of human complement (C4A and C4B) have been associated with various diseases. High-throughput methods for analysing C4 CNV are available, but they commonly do not detect the most common C4A mutation, a silencing CT insertion (CTins) leading to low protein levels. We developed a SYBR® Green labelled real-time quantitative polymerase chain reaction (qPCR) with a novel concentration range approach to address C4 CNV and deficiencies due to CTins. This method was validated in three sample sets and applied to over 1600 patient samples. CTins caused C4A deficiency in more than 70% (76/105) of the carriers. Twenty per cent (76/381) of patients with a C4A deficiency would have been erroneously recorded as having none, if the CTins had not been assessed. C4A deficiency was more common in patients than a healthy reference population, (OR = 1.60, 95%CI = 1.02-2.52, p = 0.039). The number of functional C4 genes can be straightforwardly analyzed by real-time qPCR, also with SYBR® Green labelling. Determination of CTins increases the frequency of C4A deficiency and thus helps to elucidate the genotypic versus phenotypic disease associations.
Conflict of interest statement
Figures
References
-
- Samano ES, Ribeiro Lde M, Gorescu RG, Rocha KC, Grumach AS. Involvement of C4 allotypes in the pathogenesis of human diseases. Rev Hosp Clin Fac Med Sao Paulo. 2004;59(3):138–144. - PubMed
-
- Szilagyi A, Fust G. Diseases associated with the low copy number of the C4B gene encoding C4, the fourth component of complement. Cytogenet Genome Res. 2008;123(1–4):118–130. - PubMed
-
- Mougey R. A review of the Chido/Rodgers blood group. Immunohematology. 2010;26(1):30–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

