C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts
- PMID: 22738724
- PMCID: PMC3410639
- DOI: 10.1016/j.cell.2012.06.016
C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts
Abstract
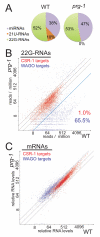
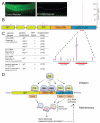
Piwi Argonautes and Piwi-interacting RNAs (piRNAs) mediate genome defense by targeting transposons. However, many piRNA species lack obvious sequence complementarity to transposons or other loci; only one C. elegans transposon is a known piRNA target. Here, we show that, in mutants lacking the Piwi Argonaute PRG-1 (and consequently its associated piRNAs/21U-RNAs), many silent loci in the germline exhibit increased levels of mRNA expression with a concomitant depletion of RNA-dependent RNA polymerase (RdRP)-derived secondary small RNAs termed 22G-RNAs. Sequences depleted of 22G-RNAs are proximal to potential target sites that base pair imperfectly but extensively to 21U-RNAs. We show that PRG-1 is required to initiate, but not to maintain, silencing of transgenes engineered to contain complementarity to endogenous 21U-RNAs. Our findings support a model in which C. elegans piRNAs utilize their enormous repertoire of targeting capacity to scan the germline transcriptome for foreign sequences, while endogenous germline-expressed genes are actively protected from piRNA-induced silencing.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


Comment in
-
Small RNAs: piRNA surveillance in the C. elegans germline.Nat Rev Genet. 2012 Jul 10;13(8):518-9. doi: 10.1038/nrg3289. Nat Rev Genet. 2012. PMID: 22777129 No abstract available.
-
Small RNAs: transmitting silence through generations.Nat Rev Mol Cell Biol. 2012 Jul 23;13(8):477. doi: 10.1038/nrm3406. Nat Rev Mol Cell Biol. 2012. PMID: 22820881 No abstract available.
References
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
-
- Aravin AA, Hannon GJ, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

