Cell-permeable, small-molecule activators of the insulin-degrading enzyme
- PMID: 22740246
- PMCID: PMC5901702
- DOI: 10.1177/1087057112451921
Cell-permeable, small-molecule activators of the insulin-degrading enzyme
Abstract
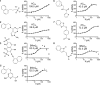
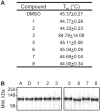
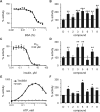
The insulin-degrading enzyme (IDE) cleaves numerous small peptides, including biologically active hormones and disease-related peptides. The propensity of IDE to degrade neurotoxic Aβ peptides marks IDE as a potential therapeutic target for Alzheimer disease. Using a synthetic reporter based on the yeast a-factor mating pheromone precursor, which is cleaved by multiple IDE orthologs, we identified seven small molecules that stimulate rat IDE activity in vitro. Half-maximal activation of IDE by the compounds is observed in vitro in the range of 43 to 198 µM. All compounds decrease the K(m) of IDE. Four compounds activate IDE in the presence of the competing substrate insulin, which disproportionately inhibits IDE activity. Two compounds stimulate rat IDE activity in a cell-based assay, indicating that they are cell permeable. The compounds demonstrate specificity for rat IDE since they do not enhance the activities of IDE orthologs, including human IDE, and they appear specific for a-factor-based reporters since they do not enhance rat IDE-mediated cleavage of Aβ-based reporters. Our results suggest that IDE activators function in the context of specific enzyme-substrate pairs, indicating that the choice of substrate must be considered in addition to target validation in IDE activator screens.
Conflict of interest statement
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Figures



References
-
- Haass C, Selkoe DJ. Soluble Protein Oligomers in Neurodegeneration: Lessons from the Alzheimer’s Amyloid Beta-Peptide. Nat. Rev. Mol. Cell. Biol. 2007;8(2):101–112. - PubMed
-
- Nalivaeva NN, Beckett C, Belyaev ND, Turner AJ. Are Amyloid-Degrading Enzymes Viable Therapeutic Targets in Alzheimer’s Disease? J. Neurochem. 2012;120(Suppl. 1):167–185. - PubMed
-
- Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-Degrading Enzyme Regulates the Levels of Insulin, Amyloid Beta-Protein, and the Beta-Amyloid Precursor Protein Intracellular Domain In Vivo. Proc. Natl. Acad. Sci. U. S. A. 2003;100(7):4162–4167. - PMC - PubMed
-
- Leissring MA, Farris W, Chang AY, Walsh DM, Wu X, Sun X, Frosch MP, Selkoe DJ. Enhanced Proteolysis of Beta-Amyloid in APP Transgenic Mice Prevents Plaque Formation, Secondary Pathology, and Premature Death. Neuron. 2003;40(6):1087–1093. - PubMed
-
- Kim M, Hersh LB, Leissring MA, Ingelsson M, Matsui T, Farris W, Lu A, Hyman BT, Selkoe DJ, Bertram L, Tanzi RE. Decreased Catalytic Activity of the Insulin Degrading Enzyme in Chromosome 10-Linked Alzheimer’s Disease Families. J. Biol. Chem. 2007;282(11):7825–7832. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

