DNA hypermethylation of alternatively spliced and repeat sequences in humans
- PMID: 22740315
- PMCID: PMC3407362
- DOI: 10.1007/s00438-012-0703-y
DNA hypermethylation of alternatively spliced and repeat sequences in humans
Abstract
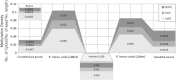
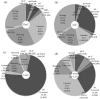
DNA methylation is presently accepted as a tentative regulatory parameter in splicing. Recently, we reported significant methylation differences among various exonic splicing-enhancing elements and alternative splicing events, based on CpG methylation data from the Human Epigenome Project for chromosomes 6, 20 and 22. Presently, using a different computational approach and the same database, we report: (a) significant increase of hypermethylation in intronic and exonic sequences close to acceptor sites, relative to overall introns and exons, respectively (1,973 CpGs examined); (b) frequent CpGs, mostly hypomethylated, in donors and infrequent CpGs mostly hypermethylated, in acceptors; and (c) hypermethylation in cassette exons which are occasionally spliced and have weaker average splicing potential, relative to constitutive exons (p < 0.0001). CpGs are hypomethylated in non-coding exons (only 16 % hypermethylation). Single-exon genes, similarly to first exons, frequently contain hypomethylated CpGs, while in internal and last exons CpGs are more frequently hypermethylated. Methylation is also more frequent in strange introns and splice sites processed by the minor spliceosome, e.g., ATAC, (p < 0.0001 in all cases), but not in sites of incomplete processing, e.g., retained introns or bleeding exons, (p = 0.706 and p = 0.313, respectively). Most Alus, which are known to contribute to transcript presentation, are heavily methylated, in contrast with other Alus, e.g., AluJo and mammalian interspersed repetitive elements which have been previously associated with alternative expression. These results elucidate the role of intragenic methylation in association with alternative splicing and facilitate the evaluation of genomic variations/polymorphisms and the development of tools for the prediction of alternative splicing events.
Figures






Similar articles
-
Intronic CpG content and alternative splicing in human genes containing a single cassette exon.Epigenetics. 2008 Mar-Apr;3(2):69-73. doi: 10.4161/epi.3.2.6066. Epub 2008 Apr 8. Epigenetics. 2008. PMID: 18418084
-
Human epigenome data reveal increased CpG methylation in alternatively spliced sites and putative exonic splicing enhancers.DNA Cell Biol. 2011 May;30(5):267-75. doi: 10.1089/dna.2010.1094. DNA Cell Biol. 2011. PMID: 21545276
-
A systematic analysis of intronic sequences downstream of 5' splice sites reveals a widespread role for U-rich motifs and TIA1/TIAL1 proteins in alternative splicing regulation.Genome Res. 2008 Aug;18(8):1247-58. doi: 10.1101/gr.073155.107. Epub 2008 May 2. Genome Res. 2008. PMID: 18456862 Free PMC article.
-
Exonization of transposed elements: A challenge and opportunity for evolution.Biochimie. 2011 Nov;93(11):1928-34. doi: 10.1016/j.biochi.2011.07.014. Epub 2011 Jul 26. Biochimie. 2011. PMID: 21787833 Review.
-
How prevalent is functional alternative splicing in the human genome?Trends Genet. 2004 Feb;20(2):68-71. doi: 10.1016/j.tig.2003.12.004. Trends Genet. 2004. PMID: 14746986 Review.
Cited by
-
The genetic basis for individual differences in mRNA splicing and APOBEC1 editing activity in murine macrophages.Genome Res. 2014 Mar;24(3):377-89. doi: 10.1101/gr.166033.113. Epub 2013 Nov 18. Genome Res. 2014. PMID: 24249727 Free PMC article.
-
Biased gene expression in early honeybee larval development.BMC Genomics. 2013 Dec 19;14:903. doi: 10.1186/1471-2164-14-903. BMC Genomics. 2013. PMID: 24350621 Free PMC article.
-
Arginine CGA codons as a source of nonsense mutations: a possible role in multivariant gene expression, control of mRNA quality, and aging.Mol Genet Genomics. 2017 Oct;292(5):1013-1026. doi: 10.1007/s00438-017-1328-y. Epub 2017 May 18. Mol Genet Genomics. 2017. PMID: 28523359
-
Oral contraceptives modify the effect of GATA3 polymorphisms on the risk of asthma at the age of 18 years via DNA methylation.Clin Epigenetics. 2014 Sep 19;6(1):17. doi: 10.1186/1868-7083-6-17. eCollection 2014. Clin Epigenetics. 2014. PMID: 25250096 Free PMC article.
-
Integrative analysis of tissue-specific methylation and alternative splicing identifies conserved transcription factor binding motifs.Nucleic Acids Res. 2013 Oct;41(18):8503-14. doi: 10.1093/nar/gkt652. Epub 2013 Jul 24. Nucleic Acids Res. 2013. PMID: 23887936 Free PMC article.
References
-
- Chodavarapu RK, Feng S, Bernatavichute YV, Chen PY, Stroud H, Yu Y, Hetzel JA, Kuo F, Kim J, Cokus SJ, Casero D, Bernal M, Huijser P, Clark AT, Kramer U, Merchant SS, Zhang X, Jacobsen SE, Pellegrini M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466(7304):388–392. doi: 10.1038/nature09147. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

