Negative elongation factor-mediated suppression of RNA polymerase II elongation of Kaposi's sarcoma-associated herpesvirus lytic gene expression
- PMID: 22740393
- PMCID: PMC3446579
- DOI: 10.1128/JVI.01012-12
Negative elongation factor-mediated suppression of RNA polymerase II elongation of Kaposi's sarcoma-associated herpesvirus lytic gene expression
Abstract
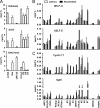
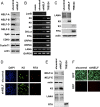
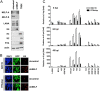
Genome-wide chromatin immunoprecipitation assays indicate that the promoter-proximal pausing of RNA polymerase II (RNAPII) is an important postinitiation step for gene regulation. During latent infection, the majority of Kaposi's sarcoma-associated herpesvirus (KSHV) genes is silenced via repressive histone marks on their promoters. Despite the absence of their expression during latency, however, several lytic promoters are enriched with activating histone marks, suggesting that mechanisms other than heterochromatin-mediated suppression contribute to preventing lytic gene expression. Here, we show that the RNAPII-mediated transcription of the KSHV OriLytL, K5, K6, and K7 (OriLytL-K7) lytic genes is paused at the elongation step during latency. Specifically, the RNAPII-mediated transcription is stalled by the host's negative elongation factor (NELF) at the promoter regions of OriLytL-K7 lytic genes during latency, leading to the hyperphosphorylation of the serine 5 residue and the hypophosphorylation of the serine 2 of the C-terminal domain of the RNAPII large subunit, a hallmark of stalled RNAPII. Consequently, depletion of NELF expression induced transition of stalled RNAPII into a productive transcription elongation at the promoter-proximal regions of OriLytL-K7 lytic genes, leading to their RTA-independent expression. Using an RTA-deficient recombinant KSHV, we also showed that expression of the K5, K6, and K7 lytic genes was highly inducible upon external stimuli compared to other lytic genes that lack RNAPII on their promoters during latency. These results indicate that the transcription elongation of KSHV OriLytL-K7 lytic genes is inhibited by NELF during latency, but can also be promptly reactivated in an RTA-independent manner upon external stimuli.
Figures




References
-
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. 2001. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol. Cell 8:327–337 - PubMed
-
- Bernstein BE, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125:315–326 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

