Drosophila mutants show NMD pathway activity is reduced, but not eliminated, in the absence of Smg6
- PMID: 22740637
- PMCID: PMC3404369
- DOI: 10.1261/rna.032821.112
Drosophila mutants show NMD pathway activity is reduced, but not eliminated, in the absence of Smg6
Abstract
The nonsense-mediated mRNA decay (NMD) pathway is best known for targeting mutant mRNAs containing premature termination codons for rapid degradation, but it is also required for regulation of many endogenous transcripts. Components of the NMD pathway were originally identified by forward genetic screens in yeast and Caenorhabditis elegans. In other organisms, the NMD pathway has been investigated by studying the homologs of these genes. We present here the first unbiased genetic screen in Drosophila designed specifically to identify genes involved in NMD. By using a highly efficient genetic mosaic approach, we have screened ∼40% of the Drosophila genome and isolated more than 40 alleles of genes required for NMD. We focus on alleles we have obtained in two known NMD components: Upf2 and Smg6. Our analysis of multiple alleles of the core NMD component Upf2 reveals that the Upf2 requirement in NMD may be separate from its requirement for viability, indicating additional critical cellular roles for this protein. Our alleles of Smg6 are the first point mutations obtained in Drosophila, and we find that Smg6 has both endonucleolytic and nonendonucleolytic roles in NMD. Thus, our genetic screens have revealed that Drosophila NMD factors play distinct roles in target regulation, similar to what is found in mammals, but distinct from the relatively similar requirements for NMD genes observed in C. elegans and yeast.
Figures
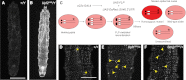

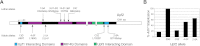
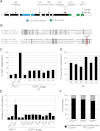
References
-
- Abràmoff MD, Magalhães PJ, Ram SJ 2004. Image processing with ImageJ. Biophotonics international 11: 36–42
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
-
- Aronoff R, Baran R, Hodgkin J 2001. Molecular identification of smg-4, required for mRNA surveillance in C. elegans. Gene 268: 153–164 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
