Disease-linked microRNA-21 exhibits drastically reduced mRNA binding and silencing activity in healthy mouse liver
- PMID: 22740638
- PMCID: PMC3404372
- DOI: 10.1261/rna.033308.112
Disease-linked microRNA-21 exhibits drastically reduced mRNA binding and silencing activity in healthy mouse liver
Abstract
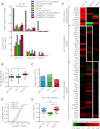
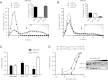
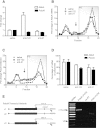
MicroRNAs (miRNAs) bind to mRNAs and fine-tune protein output by affecting mRNA stability and/or translation. miR-21 is a ubiquitous, highly abundant, and stress-responsive miRNA linked to several diseases, including cancer, fibrosis, and inflammation. Although the RNA silencing activity of miR-21 in diseased cells has been well documented, the roles of miR-21 under healthy cellular conditions are not well understood. Here, we show that pharmacological inhibition or genetic deletion of miR-21 in healthy mouse liver has little impact on regulation of canonical seed-matched mRNAs and only a limited number of genes enriched in stress response pathways. These surprisingly weak and selective regulatory effects on known and predicted target mRNAs contrast with those of other abundant liver miRNAs such as miR-122 and let-7. Moreover, miR-21 shows greatly reduced binding to polysome-associated target mRNAs compared to miR-122 and let-7. Bioinformatic analysis suggests that reduced thermodynamic stability of seed pairing and target binding may contribute to this deficiency of miR-21. Significantly, these trends are reversed in human cervical carcinoma (HeLa) cells, where miRNAs including miR-21 show enhanced target binding within polysomes and where miR-21 triggers strong degradative activity toward target mRNAs. Taken together, our results suggest that, under normal cellular conditions in liver, miR-21 activity is maintained below a threshold required for binding and silencing most of its targets. Consequently, enhanced association with polysome-associated mRNA is likely to explain in part the gain of miR-21 function often found in diseased or stressed cells.
Figures




References
-
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W 2006. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 125: 1111–1124 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
