MyoD regulates p57kip2 expression by interacting with a distant cis-element and modifying a higher order chromatin structure
- PMID: 22740650
- PMCID: PMC3458561
- DOI: 10.1093/nar/gks619
MyoD regulates p57kip2 expression by interacting with a distant cis-element and modifying a higher order chromatin structure
Abstract
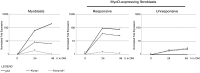
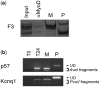
The bHLH transcription factor MyoD, the prototypical master regulator of differentiation, directs a complex program of gene expression during skeletal myogenesis. The up-regulation of the cdk inhibitor p57kip2 plays a critical role in coordinating differentiation and growth arrest during muscle development, as well as in other tissues. p57kip2 displays a highly specific expression pattern and is subject to a complex epigenetic control driving the imprinting of the paternal allele. However, the regulatory mechanisms governing its expression during development are still poorly understood. We have identified an unexpected mechanism by which MyoD regulates p57kip2 transcription in differentiating muscle cells. We show that the induction of p57kip2 requires MyoD binding to a long-distance element located within the imprinting control region KvDMR1 and the consequent release of a chromatin loop involving p57kip2 promoter. We also show that differentiation-dependent regulation of p57kip2, while involving a region implicated in the imprinting process, is distinct and hierarchically subordinated to the imprinting control. These findings highlight a novel mechanism, involving the modification of higher order chromatin structures, by which MyoD regulates gene expression. Our results also suggest that chromatin folding mediated by KvDMR1 could account for the highly restricted expression of p57kip2 during development and, possibly, for its aberrant silencing in some pathologies.
Figures



Similar articles
-
The long non-coding RNA Kcnq1ot1 controls maternal p57 expression in muscle cells by promoting H3K27me3 accumulation to an intragenic MyoD-binding region.Epigenetics Chromatin. 2019 Jan 16;12(1):8. doi: 10.1186/s13072-019-0253-1. Epigenetics Chromatin. 2019. PMID: 30651140 Free PMC article.
-
A cross-talk between DNA methylation and H3 lysine 9 dimethylation at the KvDMR1 region controls the induction of Cdkn1c in muscle cells.Epigenetics. 2016 Nov;11(11):791-803. doi: 10.1080/15592294.2016.1230576. Epub 2016 Sep 9. Epigenetics. 2016. PMID: 27611768 Free PMC article.
-
Functional interplay between MyoD and CTCF in regulating long-range chromatin interactions during differentiation.J Cell Sci. 2014 Sep 1;127(Pt 17):3757-67. doi: 10.1242/jcs.149427. Epub 2014 Jul 7. J Cell Sci. 2014. PMID: 25002401
-
Shaping Gene Expression by Landscaping Chromatin Architecture: Lessons from a Master.Mol Cell. 2018 Aug 2;71(3):375-388. doi: 10.1016/j.molcel.2018.04.025. Epub 2018 Jun 7. Mol Cell. 2018. PMID: 29887393 Free PMC article. Review.
-
MyoD and the transcriptional control of myogenesis.Semin Cell Dev Biol. 2005 Aug-Oct;16(4-5):585-95. doi: 10.1016/j.semcdb.2005.07.006. Semin Cell Dev Biol. 2005. PMID: 16099183 Review.
Cited by
-
The long non-coding RNA Kcnq1ot1 controls maternal p57 expression in muscle cells by promoting H3K27me3 accumulation to an intragenic MyoD-binding region.Epigenetics Chromatin. 2019 Jan 16;12(1):8. doi: 10.1186/s13072-019-0253-1. Epigenetics Chromatin. 2019. PMID: 30651140 Free PMC article.
-
Dlx5 and Dlx6 can antagonize cell division at the G1/S checkpoint.BMC Mol Cell Biol. 2019 Apr 11;20(1):8. doi: 10.1186/s12860-019-0191-6. BMC Mol Cell Biol. 2019. PMID: 31041891 Free PMC article.
-
Developing in 3D: the role of CTCF in cell differentiation.Development. 2018 Mar 22;145(6):dev137729. doi: 10.1242/dev.137729. Development. 2018. PMID: 29567640 Free PMC article. Review.
-
MyoD-Induced Trans-Differentiation: A Paradigm for Dissecting the Molecular Mechanisms of Cell Commitment, Differentiation and Reprogramming.Cells. 2022 Oct 31;11(21):3435. doi: 10.3390/cells11213435. Cells. 2022. PMID: 36359831 Free PMC article. Review.
-
Functional Versatility of the CDK Inhibitor p57Kip2.Front Cell Dev Biol. 2020 Oct 7;8:584590. doi: 10.3389/fcell.2020.584590. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33117811 Free PMC article. Review.
References
-
- Weintraub H. The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell. 1993;75:1241–1244. - PubMed
-
- Maione R, Amati P. Interdependence between muscle differentiation and cell-cycle control. Biochim. Biophys. Acta. 1997;1332:M19–M30. - PubMed
-
- Puri PL, Sartorelli V. Regulation of muscle regulatory factors by DNA-binding, interacting proteins, and post-transcriptional modifications. J. Cell. Physiol. 2000;185:155–173. - PubMed
-
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 2002;9:587–600. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

