X-linked sideroblastic anemia due to carboxyl-terminal ALAS2 mutations that cause loss of binding to the β-subunit of succinyl-CoA synthetase (SUCLA2)
- PMID: 22740690
- PMCID: PMC3436539
- DOI: 10.1074/jbc.M111.306423
X-linked sideroblastic anemia due to carboxyl-terminal ALAS2 mutations that cause loss of binding to the β-subunit of succinyl-CoA synthetase (SUCLA2)
Abstract
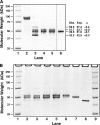
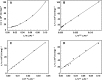
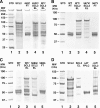
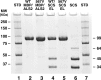
Mutations in the erythroid-specific aminolevulinic acid synthase gene (ALAS2) cause X-linked sideroblastic anemia (XLSA) by reducing mitochondrial enzymatic activity. Surprisingly, a patient with the classic XLSA phenotype had a novel exon 11 mutation encoding a recombinant enzyme (p.Met567Val) with normal activity, kinetics, and stability. Similarly, both an expressed adjacent XLSA mutation, p.Ser568Gly, and a mutation (p.Phe557Ter) lacking the 31 carboxyl-terminal residues also had normal or enhanced activity, kinetics, and stability. Because ALAS2 binds to the β subunit of succinyl-CoA synthetase (SUCLA2), the mutant proteins were tested for their ability to bind to this protein. Wild type ALAS2 bound strongly to a SUCLA2 affinity column, but the adjacent XLSA mutant enzymes and the truncated mutant did not bind. In contrast, vitamin B6-responsive XLSA mutations p.Arg452Cys and p.Arg452His, with normal in vitro enzyme activity and stability, did not interfere with binding to SUCLA2 but instead had loss of positive cooperativity for succinyl-CoA binding, an increased K(m) for succinyl-CoA, and reduced vitamin B6 affinity. Consistent with the association of SUCLA2 binding with in vivo ALAS2 activity, the p.Met567GlufsX2 mutant protein that causes X-linked protoporphyria bound strongly to SUCLA2, highlighting the probable role of an ALAS2-succinyl-CoA synthetase complex in the regulation of erythroid heme biosynthesis.
Figures


References
-
- Bishop D. F., Henderson A. S., Astrin K. H. (1990) Human δ-aminolevulinate synthase. Assignment of the housekeeping gene to 3p21 and the erythroid-specific gene to the X chromosome. Genomics 7, 207–214 - PubMed
-
- Anderson K. E., Sassa S., Bishop D. F., Desnick R. J. (2001) Disorders of heme biosynthesis. X-linked sideroblastic anemia and the porphyrias. in The Metabolic and Molecular Bases of Inherited Disease, 8th Ed. (Scriver C. R., Beaudet A. L., Sly W. S., Valle D., Childs B., Kinzler K. W., Vogelstein B., eds) pp. 2991–3062, McGraw-Hill, New York
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

