Alternative splicing in plants--coming of age
- PMID: 22743067
- PMCID: PMC3466422
- DOI: 10.1016/j.tplants.2012.06.001
Alternative splicing in plants--coming of age
Abstract
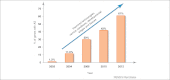
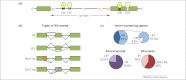
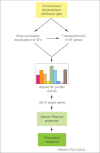
More than 60% of intron-containing genes undergo alternative splicing (AS) in plants. This number will increase when AS in different tissues, developmental stages, and environmental conditions are explored. Although the functional impact of AS on protein complexity is still understudied in plants, recent examples demonstrate its importance in regulating plant processes. AS also regulates transcript levels and the link with nonsense-mediated decay and generation of unproductive mRNAs illustrate the need for both transcriptional and AS data in gene expression analyses. AS has influenced the evolution of the complex networks of regulation of gene expression and variation in AS contributed to adaptation of plants to their environment and therefore will impact strategies for improving plant and crop phenotypes.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
References
-
- Black D.L. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 2003;72:291–336. - PubMed
-
- Graveley B.R. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. - PubMed
-
- Lareau L.F. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. - PubMed
-
- Stamm S. Function of alternative splicing. Gene. 2005;344:1–20. - PubMed
-
- Pan Q. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

