GO-Elite: a flexible solution for pathway and ontology over-representation
- PMID: 22743224
- PMCID: PMC3413395
- DOI: 10.1093/bioinformatics/bts366
GO-Elite: a flexible solution for pathway and ontology over-representation
Abstract
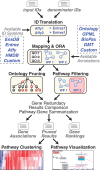
We introduce GO-Elite, a flexible and powerful pathway analysis tool for a wide array of species, identifiers (IDs), pathways, ontologies and gene sets. In addition to the Gene Ontology (GO), GO-Elite allows the user to perform over-representation analysis on any structured ontology annotations, pathway database or biological IDs (e.g. gene, protein or metabolite). GO-Elite exploits the structured nature of biological ontologies to report a minimal set of non-overlapping terms. The results can be visualized on WikiPathways or as networks. Built-in support is provided for over 60 species and 50 ID systems, covering gene, disease and phenotype ontologies, multiple pathway databases, biomarkers, and transcription factor and microRNA targets. GO-Elite is available as a web interface, GenMAPP-CS plugin and as a cross-platform application.
Availability: http://www.genmapp.org/go_elite
Figures
References
-
- Hochstenbach K., et al. Transcriptomic profile indicative of immunotoxic exposure: in vitro studies in peripheral blood mononuclear cells. Toxicol. Sci. 2010;118:19–30. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

