TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination
- PMID: 22745133
- PMCID: PMC3436586
- DOI: 10.1074/jbc.M112.362608
TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination
Abstract
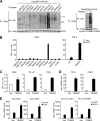
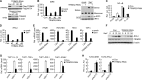
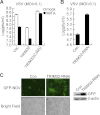
Viral infection activates several transcription factors including NF-κB and IRF3, which collaborate to induce type I interferons (IFNs) and innate antiviral response. MITA (also called STING) is a critical adaptor protein that links virus-sensing receptors to IRF3 activation upon infection by both RNA and DNA pathogens. Here we show that the E3 ubiquitin ligase tripartite motif protein 32 (TRIM32) ubiquitinated MITA and dramatically enhanced MITA-mediated induction of IFN-β. Overexpression of TRIM32 potentiated virus-triggered IFNB1 expression and cellular antiviral response. Consistently, knockdown of TRIM32 had opposite effects. TRIM32 interacted with MITA, and was located at the mitochondria and endoplasmic reticulum. TRIM32 targeted MITA for K63-linked ubiquitination at K20/150/224/236 through its E3 ubiquitin ligase activity, which promoted the interaction of MITA with TBK1. These findings suggest that TRIM32 is an important regulatory protein for innate immunity against both RNA and DNA viruses by targeting MITA for K63-linked ubiquitination and downstream activation.
Figures


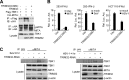
References
-
- Takeuchi O., Akira S. (2010) Pattern recognition receptors and inflammation. Cell 140, 805–820 - PubMed
-
- Akira S., Uematsu S., Takeuchi O. (2006) Pathogen recognition and innate immunity. Cell 124, 783–801 - PubMed
-
- Durbin J. E., Fernandez-Sesma A., Lee C. K., Rao T. D., Frey A. B., Moran T. M., Vukmanovic S., García-Sastre A., Levy D. E. (2000) Type I IFN modulates innate and specific antiviral immunity. J. Immunol. 164, 4220–4228 - PubMed
-
- Levy D. E., García-Sastre A. (2001) The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12, 143–156 - PubMed
-
- Levy D. E., Marié I. J. (2004) RIGging an antiviral defense–it's in the CARDs. Nat Immunol 5, 699–701 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

