Evolution of the RALF Gene Family in Plants: Gene Duplication and Selection Patterns
- PMID: 22745530
- PMCID: PMC3382376
- DOI: 10.4137/EBO.S9652
Evolution of the RALF Gene Family in Plants: Gene Duplication and Selection Patterns
Abstract
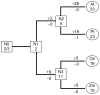
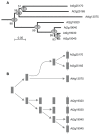
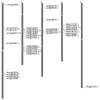
Rapid alkalinization factors (RALFs) are plant small peptides that could induce a rapid pH increase in the medium of plant cell suspension culture and play a critical role in plant development. The evolutionary process of the RALF gene family remains unclear. To obtain details of the phylogeny of these genes, this study characterized RALF genes in Arabidopsis, rice, poplar and maize. Phylogenetic trees, evolutionary patterns and molecular evolutionary rates were used to elucidate the evolutionary process of this gene family. In addition, the different signatures of selection, expression patterns, and subcellular localization of RALFs were also analyzed. We found that the RALF gene family had a rapid birth process after the separation of the eudicot and monocot species about 145 million years ago, that tandem duplication played a dominant role in the expansion of Arabidopsis and rice RALF gene family, and that RALFs were under purifying selection according to estimations of the substitution rates of these genes. We also identified a diverse expression pattern of RALF genes and predominant extracellular localization feature of RALF proteins. Our findings shed light on several key differences in RALF gene family evolution among the plant species, which may provide a scaffold for future functional analysis of this family.
Keywords: RALF; evolution; selection; tandem duplication.
Figures


References
LinkOut - more resources
Full Text Sources

