The Distribution of eIF4E-Family Members across Insecta
- PMID: 22745595
- PMCID: PMC3382400
- DOI: 10.1155/2012/960420
The Distribution of eIF4E-Family Members across Insecta
Abstract
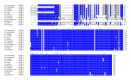
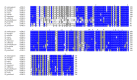
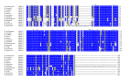
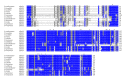
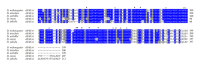
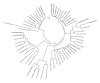
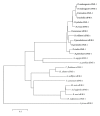
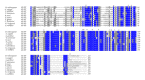
Insects are part of the earliest faunas that invaded terrestrial environments and are the first organisms that evolved controlled flight. Nowadays, insects are the most diverse animal group on the planet and comprise the majority of extant animal species described. Moreover, they have a huge impact in the biosphere as well as in all aspects of human life and economy; therefore understanding all aspects of insect biology is of great importance. In insects, as in all cells, translation is a fundamental process for gene expression. However, translation in insects has been mostly studied only in the model organism Drosophila melanogaster. We used all publicly available genomic sequences to investigate in insects the distribution of the genes encoding the cap-binding protein eIF4E, a protein that plays a crucial role in eukaryotic translation. We found that there is a diversity of multiple ortholog genes encoding eIF4E isoforms within the genus Drosophila. In striking contrast, insects outside this genus contain only a single eIF4E gene, related to D. melanogaster eIF4E-1. We also found that all insect species here analyzed contain only one Class II gene, termed 4E-HP. We discuss the possible evolutionary causes originating the multiplicity of eIF4E genes within the genus Drosophila.
Figures





Similar articles
-
Diverse cap-binding properties of Drosophila eIF4E isoforms.Biochim Biophys Acta. 2016 Oct;1864(10):1292-303. doi: 10.1016/j.bbapap.2016.06.015. Epub 2016 Jun 29. Biochim Biophys Acta. 2016. PMID: 27374989
-
Functional analysis of seven genes encoding eight translation initiation factor 4E (eIF4E) isoforms in Drosophila.Mech Dev. 2005 Apr;122(4):529-43. doi: 10.1016/j.mod.2004.11.011. Epub 2004 Dec 8. Mech Dev. 2005. PMID: 15804566
-
The alveolate translation initiation factor 4E family reveals a custom toolkit for translational control in core dinoflagellates.BMC Evol Biol. 2015 Feb 10;15(1):14. doi: 10.1186/s12862-015-0301-9. BMC Evol Biol. 2015. PMID: 25886308 Free PMC article.
-
Does phosphorylation of the cap-binding protein eIF4E play a role in translation initiation?Eur J Biochem. 2002 Nov;269(22):5350-9. doi: 10.1046/j.1432-1033.2002.03291.x. Eur J Biochem. 2002. PMID: 12423333 Free PMC article. Review.
-
The Role of the Eukaryotic Translation Initiation Factor 4E (eIF4E) in Neuropsychiatric Disorders.Front Genet. 2018 Nov 23;9:561. doi: 10.3389/fgene.2018.00561. eCollection 2018. Front Genet. 2018. PMID: 30532767 Free PMC article. Review.
Cited by
-
Eukaryotic initiation factor 4E-3 is essential for meiotic chromosome segregation, cytokinesis and male fertility in Drosophila.Development. 2012 Sep;139(17):3211-20. doi: 10.1242/dev.073122. Epub 2012 Jul 25. Development. 2012. PMID: 22833128 Free PMC article.
-
Regulation of Germ Cell mRNPs by eIF4E:4EIP Complexes: Multiple Mechanisms, One Goal.Front Cell Dev Biol. 2020 Jul 7;8:562. doi: 10.3389/fcell.2020.00562. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32733883 Free PMC article. Review.
-
Biological functions and research progress of eIF4E.Front Oncol. 2023 Aug 3;13:1076855. doi: 10.3389/fonc.2023.1076855. eCollection 2023. Front Oncol. 2023. PMID: 37601696 Free PMC article. Review.
-
Two Arabidopsis loci encode novel eukaryotic initiation factor 4E isoforms that are functionally distinct from the conserved plant eukaryotic initiation factor 4E.Plant Physiol. 2014 Apr;164(4):1820-30. doi: 10.1104/pp.113.227785. Epub 2014 Feb 5. Plant Physiol. 2014. PMID: 24501003 Free PMC article.
-
eIF4E and Interactors from Unicellular Eukaryotes.Int J Mol Sci. 2020 Mar 21;21(6):2170. doi: 10.3390/ijms21062170. Int J Mol Sci. 2020. PMID: 32245232 Free PMC article. Review.
References
-
- Mayhew PJ. Why are there so many insect species? Perspectives from fossils and phylogenies. Biological Reviews. 2007;82(3):425–454. - PubMed
-
- Grimaldi DA, Engel MS. Evolution of the Insects. New York, NY, USA: Cambridge University Press; 2005.
-
- Bisby FA, Roskov YR, Orrell TM, Nicolson D, Paglinawan LE. Catalogue of life: 2010 annual checklist. Species 2000. 2010;23(24):33–54.
-
- Engel MS, Grimaldi DA. New light shed on the oldest insect. Nature. 2004;427(6975):627–630. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
