UV resonance Raman spectroscopy monitors polyglutamine backbone and side chain hydrogen bonding and fibrillization
- PMID: 22746095
- PMCID: PMC3415266
- DOI: 10.1021/bi300551b
UV resonance Raman spectroscopy monitors polyglutamine backbone and side chain hydrogen bonding and fibrillization
Abstract
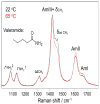
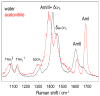
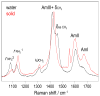
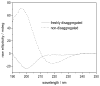
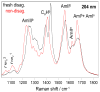
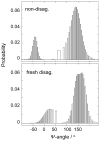
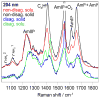
We utilize 198 and 204 nm excited UV resonance Raman spectroscopy (UVRR) and circular dichroism spectroscopy (CD) to monitor the backbone conformation and the Gln side chain hydrogen bonding (HB) of a short, mainly polyGln peptide with a D(2)Q(10)K(2) sequence (Q10). We measured the UVRR spectra of valeramide to determine the dependence of the primary amide vibrations on amide HB. We observe that a nondisaggregated Q10 (NDQ10) solution (prepared by directly dissolving the original synthesized peptide in pure water) exists in a β-sheet conformation, where the Gln side chains form hydrogen bonds to either the backbone or other Gln side chains. At 60 °C, these solutions readily form amyloid fibrils. We used the polyGln disaggregation protocol of Wetzel et al. [Wetzel, R., et al. (2006) Methods Enzymol.413, 34-74] to dissolve the Q10 β-sheet aggregates. We observe that the disaggregated Q10 (DQ10) solutions adopt PPII-like and 2.5(1)-helix conformations where the Gln side chains form hydrogen bonds with water. In contrast, these samples do not form fibrils. The NDQ10 β-sheet solution structure is essentially identical to that found in the NDQ10 solid formed upon evaporation of the solution. The DQ10 PPII and 2.5(1)-helix solution structure is essentially identical to that in the DQ10 solid. Although the NDQ10 solution readily forms fibrils when heated, the DQ10 solution does not form fibrils unless seeded with the NDQ10 solution. This result demonstrates very high activation barriers between these solution conformations. The NDQ10 fibril secondary structure is essentially identical to that of the NDQ10 solution, except that the NDQ10 fibril backbone conformational distribution is narrower than in the dissolved species. The NDQ10 fibril Gln side chain geometry is more constrained than when NDQ10 is in solution. The NDQ10 fibril structure is identical to that of the DQ10 fibril seeded by the NDQ10 solution.
Figures




Similar articles
-
UV Resonance Raman Structural Characterization of an (In)soluble Polyglutamine Peptide.J Phys Chem B. 2019 Feb 28;123(8):1749-1763. doi: 10.1021/acs.jpcb.8b10783. Epub 2019 Feb 19. J Phys Chem B. 2019. PMID: 30717595 Free PMC article.
-
Monomeric Polyglutamine Structures That Evolve into Fibrils.J Phys Chem B. 2017 Jun 22;121(24):5953-5967. doi: 10.1021/acs.jpcb.7b04060. Epub 2017 Jun 8. J Phys Chem B. 2017. PMID: 28531354
-
Polyglutamine Fibrils: New Insights into Antiparallel β-Sheet Conformational Preference and Side Chain Structure.J Phys Chem B. 2016 Mar 31;120(12):3012-26. doi: 10.1021/acs.jpcb.5b11380. Epub 2016 Mar 18. J Phys Chem B. 2016. PMID: 26947327
-
Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: a review.Analyst. 2015 Aug 7;140(15):4967-80. doi: 10.1039/c5an00342c. Analyst. 2015. PMID: 26042229 Review.
-
Determinants of the polyproline II helix from modeling studies.Adv Protein Chem. 2002;62:263-82. doi: 10.1016/s0065-3233(02)62010-8. Adv Protein Chem. 2002. PMID: 12418106 Review.
Cited by
-
Polyglutamine Solution-State Structural Propensity Is Repeat Length Dependent.J Phys Chem B. 2019 May 16;123(19):4193-4203. doi: 10.1021/acs.jpcb.9b01433. Epub 2019 May 1. J Phys Chem B. 2019. PMID: 31008597 Free PMC article.
-
Low-frequency Raman spectra of amyloid fibrils.J Chem Phys. 2025 Jun 7;162(21):215101. doi: 10.1063/5.0260500. J Chem Phys. 2025. PMID: 40459359
-
UV Resonance Raman Structural Characterization of an (In)soluble Polyglutamine Peptide.J Phys Chem B. 2019 Feb 28;123(8):1749-1763. doi: 10.1021/acs.jpcb.8b10783. Epub 2019 Feb 19. J Phys Chem B. 2019. PMID: 30717595 Free PMC article.
-
Integrative determination of the atomic structure of mutant huntingtin exon 1 fibrils implicated in Huntington's disease.bioRxiv [Preprint]. 2024 Sep 15:2023.07.21.549993. doi: 10.1101/2023.07.21.549993. bioRxiv. 2024. Update in: Nat Commun. 2024 Dec 30;15(1):10793. doi: 10.1038/s41467-024-55062-8. PMID: 37502911 Free PMC article. Updated. Preprint.
-
Polarized Raman Spectroscopy for Determining the Orientation of di-D-phenylalanine Molecules in a Nanotube.J Raman Spectrosc. 2016 Sep;47(9):1056-1062. doi: 10.1002/jrs.4884. Epub 2016 Feb 17. J Raman Spectrosc. 2016. PMID: 27795612 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

