Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes
- PMID: 22747664
- PMCID: PMC3403997
- DOI: 10.1186/1745-6150-7-20
Unproductive alternative splicing and nonsense mRNAs: a widespread phenomenon among plant circadian clock genes
Abstract
Background: Recent mapping of eukaryotic transcriptomes and spliceomes using massively parallel RNA sequencing (RNA-seq) has revealed that the extent of alternative splicing has been considerably underestimated. Evidence also suggests that many pre-mRNAs undergo unproductive alternative splicing resulting in incorporation of in-frame premature termination codons (PTCs). The destinies and potential functions of the PTC-harboring mRNAs remain poorly understood. Unproductive alternative splicing in circadian clock genes presents a special case study because the daily oscillations of protein expression levels require rapid and steep adjustments in mRNA levels.
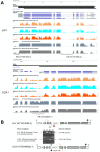
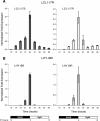
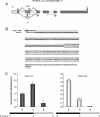
Results: We conducted a systematic survey of alternative splicing of plant circadian clock genes using RNA-seq and found that many Arabidopsis thaliana circadian clock-associated genes are alternatively spliced. Results were confirmed using reverse transcription polymerase chain reaction (RT-PCR), quantitative RT-PCR (qRT-PCR), and/or Sanger sequencing. Intron retention events were frequently observed in mRNAs of the CCA1/LHY-like subfamily of MYB transcription factors. In contrast, the REVEILLE2 (RVE2) transcript was alternatively spliced via inclusion of a "poison cassette exon" (PCE). The PCE type events introducing in-frame PTCs are conserved in some mammalian and plant serine/arginine-rich splicing factors. For some circadian genes such as CCA1 the ratio of the productive isoform (i.e., a representative splice variant encoding the full-length protein) to its PTC counterpart shifted sharply under specific environmental stress conditions.
Conclusions: Our results demonstrate that unproductive alternative splicing is a widespread phenomenon among plant circadian clock genes that frequently generates mRNA isoforms harboring in-frame PTCs. Because LHY and CCA1 are core components of the plant central circadian oscillator, the conservation of alternatively spliced variants between CCA1 and LHY and for CCA1 across phyla [2] indicates a potential role of nonsense transcripts in regulation of circadian rhythms. Most of the alternatively spliced isoforms harbor in-frame PTCs that arise from full or partial intron retention events. However, a PTC in the RVE2 transcript is introduced through a PCE event. The conservation of AS events and modulation of the relative abundance of nonsense isoforms by environmental and diurnal conditions suggests possible regulatory roles for these alternatively spliced transcripts in circadian clock function. The temperature-dependent expression of the PTC transcripts among members of CCA1/LHY subfamily indicates that alternative splicing may be involved in regulation of the clock temperature compensation mechanism.
Figures


References
-
- Filichkin SA, Breton G, Priest HD, Dharmawardhana P, Jaiswal P, Fox SE, Chory J, Michael TP, Kay S, Mockler TC. Global profiling of rice and poplar transcriptomes highlights key conserved circadian-controlled pathways and cis-regulatory modules. PLoS ONE. 2011;6:e16907. doi: 10.1371/journal.pone.0016907. - DOI - PMC - PubMed
-
- Cumbie JS, Kimbrel JA, Di Y, Schafer DW, Wilhelm L, Fox SE, Sullivan CM, Curzon AD, Carrington JC, Mockler TC, Chang JH. GENE-counter: a computational pipeline for the analysis of RNA-Seq data for gene expression differences. PLoS ONE. 2011;6:e25279. doi: 10.1371/journal.pone.0025279. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

