Transcriptome characterization via 454 pyrosequencing of the annelid Pristina leidyi, an emerging model for studying the evolution of regeneration
- PMID: 22747785
- PMCID: PMC3464666
- DOI: 10.1186/1471-2164-13-287
Transcriptome characterization via 454 pyrosequencing of the annelid Pristina leidyi, an emerging model for studying the evolution of regeneration
Abstract
Background: The naid annelids contain a number of species that vary in their ability to regenerate lost body parts, making them excellent candidates for evolution of regeneration studies. However, scant sequence data exists to facilitate such studies. We constructed a cDNA library from the naid Pristina leidyi, a species that is highly regenerative and also reproduces asexually by fission, using material from a range of regeneration and fission stages for our library. We then sequenced the transcriptome of P. leidyi using 454 technology.
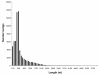
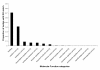
Results: 454 sequencing produced 1,550,174 reads with an average read length of 376 nucleotides. Assembly of 454 sequence reads resulted in 64,522 isogroups and 46,679 singletons for a total of 111,201 unigenes in this transcriptome. We estimate that over 95% of the transcripts in our library are present in our transcriptome. 17.7% of isogroups had significant BLAST hits to the UniProt database and these include putative homologs of a number of genes relevant to regeneration research. Although many sequences are incomplete, the mean sequence length of transcripts (isotigs) is 707 nucleotides. Thus, many sequences are large enough to be immediately useful for downstream applications such as gene expression analyses. Using in situ hybridization, we show that two Wnt/β-catenin pathway genes (homologs of frizzled and β-catenin) present in our transcriptome are expressed in the regeneration blastema of P. leidyi, demonstrating the usefulness of this resource for regeneration research.
Conclusions: 454 sequencing is a rapid and efficient approach for identifying large numbers of genes in an organism that lacks a sequenced genome. This transcriptome dataset will be a valuable resource for molecular analyses of regeneration in P. leidyi and will serve as a starting point for comparisons to non-regenerating naids. It also contributes significantly to the still limited genomic resources available for annelids and lophotrochozoans more generally.
Figures

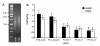



Similar articles
-
Distinct patterns of gene expression during regeneration and asexual reproduction in the annelid Pristina leidyi.J Exp Zool B Mol Dev Evol. 2022 Nov;338(7):405-420. doi: 10.1002/jez.b.23143. Epub 2022 May 23. J Exp Zool B Mol Dev Evol. 2022. PMID: 35604322 Free PMC article.
-
Journey beyond the embryo: The beauty of Pristina and naidine annelids for studying regeneration and agametic reproduction.Curr Top Dev Biol. 2022;147:469-495. doi: 10.1016/bs.ctdb.2021.12.020. Epub 2022 Mar 7. Curr Top Dev Biol. 2022. PMID: 35337459
-
Investment choices in post-embryonic development: quantifying interactions among growth, regeneration, and asexual reproduction in the annelid Pristina leidyi.J Exp Zool B Mol Dev Evol. 2013 Dec;320(8):471-88. doi: 10.1002/jez.b.22523. Epub 2013 Aug 2. J Exp Zool B Mol Dev Evol. 2013. PMID: 23913524
-
Wnt/β-Catenin Signaling Pathway Governs a Full Program for Dopaminergic Neuron Survival, Neurorescue and Regeneration in the MPTP Mouse Model of Parkinson's Disease.Int J Mol Sci. 2018 Nov 24;19(12):3743. doi: 10.3390/ijms19123743. Int J Mol Sci. 2018. PMID: 30477246 Free PMC article. Review.
-
The roles of Wnt/β-catenin pathway in tissue development and regenerative medicine.J Cell Physiol. 2018 Aug;233(8):5598-5612. doi: 10.1002/jcp.26265. Epub 2018 Mar 7. J Cell Physiol. 2018. PMID: 29150936 Review.
Cited by
-
Characterization of Liaoning cashmere goat transcriptome: sequencing, de novo assembly, functional annotation and comparative analysis.PLoS One. 2013 Oct 9;8(10):e77062. doi: 10.1371/journal.pone.0077062. eCollection 2013. PLoS One. 2013. PMID: 24130835 Free PMC article.
-
De novo assembly and characterization of the fruit transcriptome of Chinese jujube (Ziziphus jujuba Mill.) Using 454 pyrosequencing and the development of novel tri-nucleotide SSR markers.PLoS One. 2014 Sep 3;9(9):e106438. doi: 10.1371/journal.pone.0106438. eCollection 2014. PLoS One. 2014. PMID: 25184704 Free PMC article.
-
Comparative transcriptomics in Syllidae (Annelida) indicates that posterior regeneration and regular growth are comparable, while anterior regeneration is a distinct process.BMC Genomics. 2019 Nov 14;20(1):855. doi: 10.1186/s12864-019-6223-y. BMC Genomics. 2019. PMID: 31726983 Free PMC article.
-
Injury-Induced Innate Immune Response During Segment Regeneration of the Earthworm, Eisenia andrei.Int J Mol Sci. 2021 Feb 27;22(5):2363. doi: 10.3390/ijms22052363. Int J Mol Sci. 2021. PMID: 33673408 Free PMC article.
-
Annelid adult cell type diversity and their pluripotent cellular origins.Nat Commun. 2024 Apr 12;15(1):3194. doi: 10.1038/s41467-024-47401-6. Nat Commun. 2024. PMID: 38609365 Free PMC article.
References
-
- Réaumur RAF. Sur les diverses reproductions qui se font dans les Ecrevisse, les Omars, les Crabes, etc. et entr'autres sur celles de leurs Jambes et de leurs Ecailles. Mem Acad Royal Sci. 1712. pp. 223–245.
-
- Trembley A. Memoires pour servir a l'Histoire d'un Genre de Polypes d'Eau douce, a Bras en Forme de Cornes. 1744.
-
- Bely AE, Nyberg KG. Evolution of animal regeneration: re-emergence of a field. Trends Ecol Evol. 2010;25:161–170. - PubMed
-
- Goss RJ. The evolution of regeneration: adaptive or inherent? J Theor Biol. 1992;159:241–260. - PubMed
-
- Bely AE. Evolutionary loss of animal regeneration: pattern and process. Integr Comp Biol. 2010;50(4):515–527. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

