Speed control: cogs and gears that drive the circadian clock
- PMID: 22748426
- PMCID: PMC3434952
- DOI: 10.1016/j.tins.2012.05.007
Speed control: cogs and gears that drive the circadian clock
Abstract
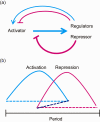
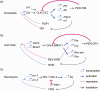
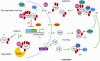
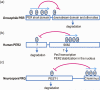
In most organisms, an intrinsic circadian (~24-h) timekeeping system drives rhythms of physiology and behavior. Within cells that contain a circadian clock, specific transcriptional activators and repressors reciprocally regulate each other to generate a basic molecular oscillator. A mismatch of the period generated by this oscillator with the external environment creates circadian disruption, which can have adverse effects on neural function. Although several clock genes have been extensively characterized, a fundamental question remains: how do these genes work together to generate a ~24-h period? Period-altering mutations in clock genes can affect any of multiple regulated steps in the molecular oscillator. In this review, we examine the regulatory mechanisms that contribute to setting the pace of the circadian oscillator.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures


References
-
- Foster RG, Roenneberg T. Human responses to the geophysical daily, annual and lunar cycles. Curr Biol. 2008;18:R784–R794. - PubMed
-
- Bechtold DA, et al. Circadian dysfunction in disease. Trends in Pharmacological Sciences. 2010;31:191–198. - PubMed
-
- Allada R, et al. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

