Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits
- PMID: 22751017
- PMCID: PMC3654374
- DOI: 10.1038/nsmb.2308
Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits
Abstract
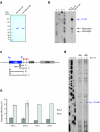
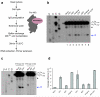
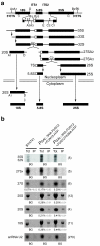
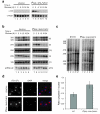
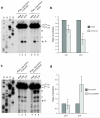
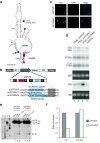
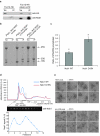
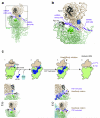
In the final steps of yeast ribosome synthesis, immature translation-incompetent pre-40S particles that contain 20S pre-rRNA are converted to the mature translation-competent subunits containing the 18S rRNA. An assay for 20S pre-rRNA cleavage in purified pre-40S particles showed that cleavage by the PIN domain endonuclease Nob1 was strongly stimulated by the GTPase activity of Fun12, the yeast homolog of cytoplasmic translation initiation factor eIF5b. Cleavage of the 20S pre-rRNA was also inhibited in vivo and in vitro by blocking binding of Fun12 to the 25S rRNA through specific methylation of its binding site. Cleavage competent pre-40S particles stably associated with Fun12 and formed 80S complexes with 60S ribosomal subunits. We propose that recruitment of 60S subunits promotes GTP hydrolysis by Fun12, leading to structural rearrangements within the pre-40S particle that bring Nob1 and the pre-rRNA cleavage site together.
Figures
Comment in
-
Getting ready to commit: ribosomes rehearse translation.Nat Struct Mol Biol. 2012 Sep;19(9):861-2. doi: 10.1038/nsmb.2368. Nat Struct Mol Biol. 2012. PMID: 22955931 No abstract available.
Similar articles
-
Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae.EMBO J. 2010 Jan 6;29(1):80-92. doi: 10.1038/emboj.2009.307. Epub 2009 Nov 5. EMBO J. 2010. PMID: 19893492 Free PMC article.
-
Immature large ribosomal subunits containing the 7S pre-rRNA can engage in translation in Saccharomyces cerevisiae.RNA Biol. 2015;12(8):838-46. doi: 10.1080/15476286.2015.1058477. RNA Biol. 2015. PMID: 26151772 Free PMC article.
-
Interaction between 25S rRNA A loop and eukaryotic translation initiation factor 5B promotes subunit joining and ensures stringent AUG selection.Mol Cell Biol. 2013 Sep;33(18):3540-8. doi: 10.1128/MCB.00771-13. Epub 2013 Jul 8. Mol Cell Biol. 2013. PMID: 23836883 Free PMC article.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Inside the 40S ribosome assembly machinery.Curr Opin Chem Biol. 2011 Oct;15(5):657-63. doi: 10.1016/j.cbpa.2011.07.023. Epub 2011 Aug 20. Curr Opin Chem Biol. 2011. PMID: 21862385 Free PMC article. Review.
Cited by
-
The large ribosomal subunit protein L9 enables the growth of EF-P deficient cells and enhances small subunit maturation.PLoS One. 2015 Apr 16;10(4):e0120060. doi: 10.1371/journal.pone.0120060. eCollection 2015. PLoS One. 2015. PMID: 25879934 Free PMC article.
-
Ribosome assembly factor Adenylate Kinase 6 maintains cell proliferation and cell size homeostasis during root growth.New Phytol. 2020 Mar;225(5):2064-2076. doi: 10.1111/nph.16291. Epub 2019 Dec 2. New Phytol. 2020. PMID: 31665812 Free PMC article.
-
Using DMS-MaPseq to uncover the roles of DEAD-box proteins in ribosome assembly.Methods. 2022 Aug;204:249-257. doi: 10.1016/j.ymeth.2022.05.001. Epub 2022 May 9. Methods. 2022. PMID: 35550176 Free PMC article.
-
Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits.Nucleic Acids Res. 2014 Oct 29;42(19):12189-99. doi: 10.1093/nar/gku878. Epub 2014 Oct 7. Nucleic Acids Res. 2014. PMID: 25294836 Free PMC article.
-
Eukaryotic ribosome assembly, transport and quality control.Nat Struct Mol Biol. 2017 Sep 7;24(9):689-699. doi: 10.1038/nsmb.3454. Nat Struct Mol Biol. 2017. PMID: 28880863 Review.
References
-
- Inoue K, Alsina J, Chen J, Inouye M. Suppression of defective ribosome assembly in a rbfA deletion mutant by overexpression of Era, an essential GTPase in Escherichia coli. Mol. Microbiol. 2003;48:1005–1016. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

