Structural diversity and African origin of the 17q21.31 inversion polymorphism
- PMID: 22751100
- PMCID: PMC3408829
- DOI: 10.1038/ng.2335
Structural diversity and African origin of the 17q21.31 inversion polymorphism
Abstract
The 17q21.31 inversion polymorphism exists either as direct (H1) or inverted (H2) haplotypes with differential predispositions to disease and selection. We investigated its genetic diversity in 2,700 individuals, with an emphasis on African populations. We characterize eight structural haplotypes due to complex rearrangements that vary in size from 1.08-1.49 Mb and provide evidence for a 30-kb H1-H2 double recombination event. We show that recurrent partial duplications of the KANSL1 gene have occurred on both the H1 and H2 haplotypes and have risen to high frequency in European populations. We identify a likely ancestral H2 haplotype (H2') lacking these duplications that is enriched among African hunter-gatherer groups yet essentially absent from West African populations. Whereas H1 and H2 segmental duplications arose independently and before human migration out of Africa, they have reached high frequencies recently among Europeans, either because of extraordinary genetic drift or selective sweeps.
Figures
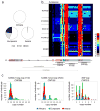
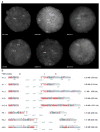


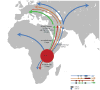
Similar articles
-
Structural haplotypes and recent evolution of the human 17q21.31 region.Nat Genet. 2012 Jul 1;44(8):881-5. doi: 10.1038/ng.2334. Nat Genet. 2012. PMID: 22751096 Free PMC article.
-
Recurrent inversion events at 17q21.31 microdeletion locus are linked to the MAPT H2 haplotype.Cytogenet Genome Res. 2010;129(4):275-9. doi: 10.1159/000315901. Epub 2010 Jul 6. Cytogenet Genome Res. 2010. PMID: 20606400 Free PMC article.
-
Reassessing the Evolutionary History of the 17q21 Inversion Polymorphism.Genome Biol Evol. 2015 Nov 11;7(12):3239-48. doi: 10.1093/gbe/evv214. Genome Biol Evol. 2015. PMID: 26560338 Free PMC article.
-
Evidence suggesting that Homo neanderthalensis contributed the H2 MAPT haplotype to Homo sapiens.Biochem Soc Trans. 2005 Aug;33(Pt 4):582-5. doi: 10.1042/BST0330582. Biochem Soc Trans. 2005. PMID: 16042549 Review.
-
From 1997 to 2007: a decade journey through the H1 haplotype on 17q21 chromosome.Parkinsonism Relat Disord. 2009 Jan;15(1):2-5. doi: 10.1016/j.parkreldis.2008.03.001. Epub 2008 Apr 18. Parkinsonism Relat Disord. 2009. PMID: 18424220 Review.
Cited by
-
The influence of 17q21.31 and APOE genetic ancestry on neurodegenerative disease risk.Front Aging Neurosci. 2022 Oct 20;14:1021918. doi: 10.3389/fnagi.2022.1021918. eCollection 2022. Front Aging Neurosci. 2022. PMID: 36337698 Free PMC article. Review.
-
Evidence for opposing selective forces operating on human-specific duplicated TCAF genes in Neanderthals and humans.Nat Commun. 2021 Aug 25;12(1):5118. doi: 10.1038/s41467-021-25435-4. Nat Commun. 2021. PMID: 34433829 Free PMC article.
-
Disease consequences of human adaptation.Appl Transl Genom. 2013 Aug 22;2:42-47. doi: 10.1016/j.atg.2013.08.001. eCollection 2013 Dec 1. Appl Transl Genom. 2013. PMID: 27896054 Free PMC article. Review.
-
Impact of APOE and MAPT genetic profile on the cognitive functions among Amyotrophic Lateral Sclerosis Tunisian patients.J Neural Transm (Vienna). 2025 Apr;132(4):609-618. doi: 10.1007/s00702-024-02870-3. Epub 2025 Jan 3. J Neural Transm (Vienna). 2025. PMID: 39751824
-
Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity.Nat Genet. 2015 Aug;47(8):921-5. doi: 10.1038/ng.3340. Epub 2015 Jun 22. Nat Genet. 2015. PMID: 26098870 Free PMC article.
References
-
- Tuzun E, et al. Fine-scale structural variation of the human genome. Nat Genet. 2005;37:727–32. - PubMed
Publication types
MeSH terms
Grants and funding
- F32HG006070/HG/NHGRI NIH HHS/United States
- T32HG00035/HG/NHGRI NIH HHS/United States
- T32HG000044/HG/NHGRI NIH HHS/United States
- F32 GM097807/GM/NIGMS NIH HHS/United States
- T32 HG000035/HG/NHGRI NIH HHS/United States
- HG002385/HG/NHGRI NIH HHS/United States
- HG004120/HG/NHGRI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- T32 HG000044/HG/NHGRI NIH HHS/United States
- F32 HG006070/HG/NHGRI NIH HHS/United States
- R01 HG002385/HG/NHGRI NIH HHS/United States
- P01 HG004120/HG/NHGRI NIH HHS/United States
- F32GM097807/GM/NIGMS NIH HHS/United States
- DP1 ES022577/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

