RET is a potential tumor suppressor gene in colorectal cancer
- PMID: 22751117
- PMCID: PMC3465636
- DOI: 10.1038/onc.2012.225
RET is a potential tumor suppressor gene in colorectal cancer
Abstract
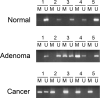
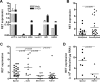
Cancer arises as the consequence of mutations and epigenetic alterations that activate oncogenes and inactivate tumor suppressor genes. Through a genome-wide screen for methylated genes in colon neoplasms, we identified aberrantly methylated RET in colorectal cancer. RET, a transmembrane receptor tyrosine kinase and a receptor for the glial cell-derived neurotrophic factor family ligands, was one of the first oncogenes to be identified, and has been shown to be an oncogene in thyroid cancer and pheochromocytoma. However, unexpectedly, we found RET is methylated in 27% of colon adenomas and in 63% of colorectal cancers, and now provide evidence that RET has tumor suppressor activity in colon cancer. The aberrant methylation of RET correlates with decreased RET expression, whereas the restoration of RET in colorectal cancer cell lines results in apoptosis. Furthermore, in support of a tumor suppressor function of RET, mutant RET has also been found in primary colorectal cancer. We now show that these mutations inactivate RET, which is consistent with RET being a tumor suppressor gene in the colon. These findings suggest that the aberrant methylation of RET and the mutational inactivation of RET promote colorectal cancer formation, and that RET can serve as a tumor suppressor gene in the colon. Moreover, the increased frequency of methylated RET in colon cancers compared with adenomas suggests RET inactivation is involved in the progression of colon adenomas to cancer.
Figures


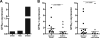
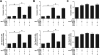
References
-
- Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer and Metastasis Reviews. 2004;23:29–39. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

