Integration of the movement of signaling microclusters with cellular motility in immunological synapses
- PMID: 22751140
- PMCID: PMC3902181
- DOI: 10.1038/ni.2364
Integration of the movement of signaling microclusters with cellular motility in immunological synapses
Abstract
Immune synapses form between T cells and antigen-presenting cells (APCs). Increasing evidence suggests synapses must form flexibly to accommodate ongoing motility and displacement of the synapse. Here, time-lapse total internal reflection fluorescence (TIRF) microscopy showed that signaling via the T cell antigen receptor (TCR) occurred during synapse translation. TCR microclusters in motile synapses did not flow directly into supramolecular activating complexes (SMACs) but were directed, independently of myosin II contractility, toward an F-actin-poor 'sink' region. Inward microcluster flow often followed collapse of the leading edge, which suggested that actin depolymerization regulated microcluster flow and the formation of SMACs. The coordination of TCR movement with the translocation of this 'sink' shows how T cells coordinate TCR signaling and microcluster flow in dynamic physiological synapses.
Figures



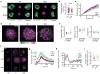

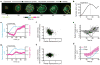

References
-
- Krummel MF, Sjaastad MD, Wülfing C, Davis MM. Differential Clustering of CD4 and CD3ζ During T Cell Recognition. Science. 2000;289:1349–1352. - PubMed
-
- Grakoui A, et al. The immunological synapse: a molecular machine controlling T cell activation. Science. 1999;285:221–227. - PubMed
-
- Wülfing C, Davis MM. A Receptor/Cytoskeletal Movement Triggered by Costimulation During T Cell Activation. Science. 1998;282:2266–2269. - PubMed
-
- Monks CRF, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395:82–86. - PubMed
-
- Yokosuka T, et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat Immunol. 2005;6:1253–1262. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

