From cis-regulatory elements to complex RNPs and back
- PMID: 22751153
- PMCID: PMC3385959
- DOI: 10.1101/cshperspect.a012245
From cis-regulatory elements to complex RNPs and back
Abstract
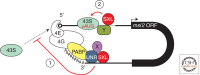
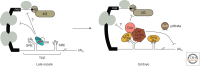
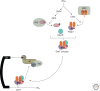
Messenger RNAs (mRNAs), the templates for translation, have evolved to harbor abundant cis-acting sequences that affect their posttranscriptional fates. These elements are frequently located in the untranslated regions and serve as binding sites for trans-acting factors, RNA-binding proteins, and/or small non-coding RNAs. This article provides a systematic synopsis of cis-acting elements, trans-acting factors, and the mechanisms by which they affect translation. It also highlights recent technical advances that have ushered in the era of transcriptome-wide studies of the ribonucleoprotein complexes formed by mRNAs and their trans-acting factors.
Figures
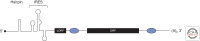

References
-
- Anderson P, Kedersha N 2009. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
