Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation
- PMID: 22751214
- PMCID: PMC3406899
- DOI: 10.1105/tpc.112.099002
Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation
Abstract
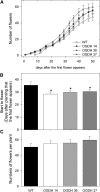
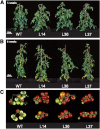
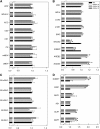
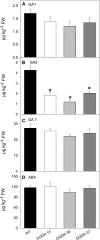
Transgenic tomato (Solanum lycopersicum) plants expressing a fragment of the gene encoding the E1 subunit of the 2-oxoglutarate dehydrogenase complex in the antisense orientation and exhibiting substantial reductions in the activity of this enzyme exhibit a considerably reduced rate of respiration. They were, however, characterized by largely unaltered photosynthetic rates and fruit yields but restricted leaf, stem, and root growth. These lines displayed markedly altered metabolic profiles, including changes in tricarboxylic acid cycle intermediates and in the majority of the amino acids but unaltered pyridine nucleotide content both in leaves and during the progression of fruit ripening. Moreover, they displayed a generally accelerated development exhibiting early flowering, accelerated fruit ripening, and a markedly earlier onset of leaf senescence. In addition, transcript and selective hormone profiling of gibberellins and abscisic acid revealed changes only in the former coupled to changes in transcripts encoding enzymes of gibberellin biosynthesis. The data obtained are discussed in the context of the importance of this enzyme in both photosynthetic and respiratory metabolism as well as in programs of plant development connected to carbon-nitrogen interactions.
Figures







Similar articles
-
Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth.Plant Physiol. 2008 May;147(1):115-27. doi: 10.1104/pp.108.117978. Epub 2008 Mar 21. Plant Physiol. 2008. PMID: 18359839 Free PMC article.
-
Mild reductions in cytosolic NADP-dependent isocitrate dehydrogenase activity result in lower amino acid contents and pigmentation without impacting growth.Amino Acids. 2010 Oct;39(4):1055-66. doi: 10.1007/s00726-010-0617-0. Epub 2010 May 16. Amino Acids. 2010. PMID: 20473773 Free PMC article.
-
Deficiency of mitochondrial fumarase activity in tomato plants impairs photosynthesis via an effect on stomatal function.Plant J. 2007 Jun;50(6):1093-106. doi: 10.1111/j.1365-313X.2007.03115.x. Epub 2007 Apr 25. Plant J. 2007. PMID: 17461782
-
The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: a review.Plant Mol Biol. 2021 Jun;106(3):223-238. doi: 10.1007/s11103-021-01128-w. Epub 2021 Feb 25. Plant Mol Biol. 2021. PMID: 33634368 Review.
-
On the role of the tricarboxylic acid cycle in plant productivity.J Integr Plant Biol. 2018 Dec;60(12):1199-1216. doi: 10.1111/jipb.12690. Epub 2018 Sep 12. J Integr Plant Biol. 2018. PMID: 29917310 Review.
Cited by
-
Genome-Wide Identification of the Aconitase Gene Family in Tomato (Solanum lycopersicum) and CRISPR-Based Functional Characterization of SlACO2 on Male-Sterility.Int J Mol Sci. 2022 Nov 12;23(22):13963. doi: 10.3390/ijms232213963. Int J Mol Sci. 2022. PMID: 36430441 Free PMC article.
-
Characterization of a NADH-dependent glutamate dehydrogenase mutant of Arabidopsis demonstrates the key role of this enzyme in root carbon and nitrogen metabolism.Plant Cell. 2012 Oct;24(10):4044-65. doi: 10.1105/tpc.112.103689. Epub 2012 Oct 9. Plant Cell. 2012. PMID: 23054470 Free PMC article.
-
Network analysis of noncoding RNAs in pepper provides insights into fruit ripening control.Sci Rep. 2019 Jun 19;9(1):8734. doi: 10.1038/s41598-019-45427-1. Sci Rep. 2019. PMID: 31217463 Free PMC article.
-
2-Oxoglutarate: linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis.Front Plant Sci. 2014 Oct 15;5:552. doi: 10.3389/fpls.2014.00552. eCollection 2014. Front Plant Sci. 2014. PMID: 25360142 Free PMC article. Review.
-
Mitochondria in photosynthetic cells: Coordinating redox control and energy balance.Plant Physiol. 2023 Apr 3;191(4):2104-2119. doi: 10.1093/plphys/kiac541. Plant Physiol. 2023. PMID: 36440979 Free PMC article.
References
-
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Larson T.R., Tohge T., Krahnert I., Witt S., Obata T., Schauer N., Graham I.A., Leaver C.J., Fernie A.R. (2010). Identification of the 2-hydroxyglutarate and isovaleryl-CoA dehydrogenases as alternative electron donors linking lysine catabolism to the electron transport chain of Arabidopsis mitochondria. Plant Cell 22: 1549–1563 - PMC - PubMed
-
- Araújo W.L., Ishizaki K., Nunes-Nesi A., Tohge T., Larson T.R., Krahnert I., Balbo I., Witt S., Dörmann P., Graham I.A., Leaver C.J., Fernie A.R. (2011b). Analysis of a range of catabolic mutants provides evidence that phytanoyl-coenzyme A does not act as a substrate of the electron-transfer flavoprotein/electron-transfer flavoprotein:ubiquinone oxidoreductase complex in Arabidopsis during dark-induced senescence. Plant Physiol. 157: 55–69 - PMC - PubMed
-
- Araújo W.L., Nunes-Nesi A., Nikoloski Z., Sweetlove L.J., Fernie A.R. (2012). Metabolic control and regulation of the tricarboxylic acid cycle in photosynthetic and heterotrophic plant tissues. Plant Cell Environ. 35: 1–21 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

